Cell-autonomous effects of APOE4 in restricting microglial response in brain homeostasis and Alzheimer's disease
- PMID: 37857825
- PMCID: PMC11980647
- DOI: 10.1038/s41590-023-01640-9
Cell-autonomous effects of APOE4 in restricting microglial response in brain homeostasis and Alzheimer's disease
Abstract
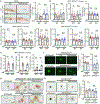
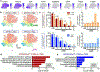
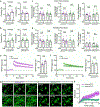
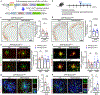
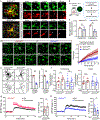
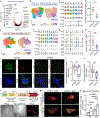
Microglial involvement in Alzheimer's disease (AD) pathology has emerged as a risk-determining pathogenic event. While apolipoprotein E (APOE) is known to modify AD risk, it remains unclear how microglial apoE impacts brain cognition and AD pathology. Here, using conditional mouse models expressing apoE isoforms in microglia and central nervous system-associated macrophages (CAMs), we demonstrate a cell-autonomous effect of apoE3-mediated microglial activation and function, which are negated by apoE4. Expression of apoE3 in microglia/CAMs improves cognitive function, increases microglia surrounding amyloid plaque and reduces amyloid pathology and associated toxicity, whereas apoE4 expression either compromises or has no effects on these outcomes by impairing lipid metabolism. Single-cell transcriptomic profiling reveals increased antigen presentation and interferon pathways upon apoE3 expression. In contrast, apoE4 expression downregulates complement and lysosomal pathways, and promotes stress-related responses. Moreover, in the presence of mouse endogenous apoE, microglial apoE4 exacerbates amyloid pathology. Finally, we observed a reduction in Lgals3-positive responsive microglia surrounding amyloid plaque and an increased accumulation of lipid droplets in APOE4 human brains and induced pluripotent stem cell-derived microglia. Our findings establish critical isoform-dependent effects of microglia/CAM-expressed apoE in brain function and the development of amyloid pathology, providing new insight into how apoE4 vastly increases AD risk.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
- AG069701/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- AG062110/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R37 AG027924/AG/NIA NIH HHS/United States
- U19 AG069701/AG/NIA NIH HHS/United States
- AG027924/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

