Integrative analyses highlight functional regulatory variants associated with neuropsychiatric diseases
- PMID: 37857935
- PMCID: PMC10859123
- DOI: 10.1038/s41588-023-01533-5
Integrative analyses highlight functional regulatory variants associated with neuropsychiatric diseases
Abstract
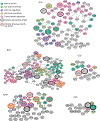
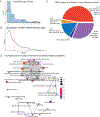
Noncoding variants of presumed regulatory function contribute to the heritability of neuropsychiatric disease. A total of 2,221 noncoding variants connected to risk for ten neuropsychiatric disorders, including autism spectrum disorder, attention deficit hyperactivity disorder, bipolar disorder, borderline personality disorder, major depression, generalized anxiety disorder, panic disorder, post-traumatic stress disorder, obsessive-compulsive disorder and schizophrenia, were studied in developing human neural cells. Integrating epigenomic and transcriptomic data with massively parallel reporter assays identified differentially-active single-nucleotide variants (daSNVs) in specific neural cell types. Expression-gene mapping, network analyses and chromatin looping nominated candidate disease-relevant target genes modulated by these daSNVs. Follow-up integration of daSNV gene editing with clinical cohort analyses suggested that magnesium transport dysfunction may increase neuropsychiatric disease risk and indicated that common genetic pathomechanisms may mediate specific symptoms that are shared across multiple neuropsychiatric diseases.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests:
Vafa Bayat and Payam Etminani are employees of Bitscopic, Inc. The remaining authors declare no competing interests.
Figures













References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

