UGDH promotes tumor-initiating cells and a fibroinflammatory tumor microenvironment in ovarian cancer
- PMID: 37858159
- PMCID: PMC10585874
- DOI: 10.1186/s13046-023-02820-z
UGDH promotes tumor-initiating cells and a fibroinflammatory tumor microenvironment in ovarian cancer
Abstract
Background: Epithelial ovarian cancer (EOC) is a global health burden, with the poorest five-year survival rate of the gynecological malignancies due to diagnosis at advanced stage and high recurrence rate. Recurrence in EOC is driven by the survival of chemoresistant, stem-like tumor-initiating cells (TICs) that are supported by a complex extracellular matrix and immunosuppressive microenvironment. To target TICs to prevent recurrence, we identified genes critical for TIC viability from a whole genome siRNA screen. A top hit was the cancer-associated, proteoglycan subunit synthesis enzyme UDP-glucose dehydrogenase (UGDH).
Methods: Immunohistochemistry was used to characterize UGDH expression in histological and molecular subtypes of EOC. EOC cell lines were subtyped according to the molecular subtypes and the functional effects of modulating UGDH expression in vitro and in vivo in C1/Mesenchymal and C4/Differentiated subtype cell lines was examined.
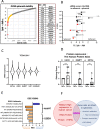
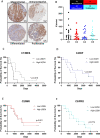
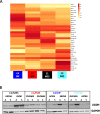
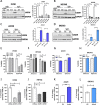
Results: High UGDH expression was observed in high-grade serous ovarian cancers and a distinctive survival prognostic for UGDH expression was revealed when serous cancers were stratified by molecular subtype. High UGDH was associated with a poor prognosis in the C1/Mesenchymal subtype and low UGDH was associated with poor prognosis in the C4/Differentiated subtype. Knockdown of UGDH in the C1/mesenchymal molecular subtype reduced spheroid formation and viability and reduced the CD133 + /ALDH high TIC population. Conversely, overexpression of UGDH in the C4/Differentiated subtype reduced the TIC population. In co-culture models, UGDH expression in spheroids affected the gene expression of mesothelial cells causing changes to matrix remodeling proteins, and fibroblast collagen production. Inflammatory cytokine expression of spheroids was altered by UGDH expression. The effect of UGDH knockdown or overexpression in the C1/ Mesenchymal and C4/Differentiated subtypes respectively was tested on mouse intrabursal xenografts and showed dynamic changes to the tumor stroma. Knockdown of UGDH improved survival and reduced tumor burden in C1/Mesenchymal compared to controls.
Conclusions: These data show that modulation of UGDH expression in ovarian cancer reveals distinct roles for UGDH in the C1/Mesenchymal and C4/Differentiated molecular subtypes of EOC, influencing the tumor microenvironmental composition. UGDH is a strong potential therapeutic target in TICs, for the treatment of EOC, particularly in patients with the mesenchymal molecular subtype.
Keywords: Mesenchymal; Molecular subtypes; Ovarian cancer; Tumor microenvironment; UGDH.
© 2023. Italian National Cancer Institute ‘Regina Elena’.
Conflict of interest statement
Authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

