A CRISPR activation screen identifies MUC-21 as critical for resistance to NK and T cell-mediated cytotoxicity
- PMID: 37858248
- PMCID: PMC10588101
- DOI: 10.1186/s13046-023-02840-9
A CRISPR activation screen identifies MUC-21 as critical for resistance to NK and T cell-mediated cytotoxicity
Abstract
Background: Immunotherapy has significantly advanced cancer treatments, but many patients do not respond to it, partly due to immunosuppressive mechanisms used by tumor cells. These cells employ immunosuppressive ligands to evade detection and elimination by the immune system. Therefore, the discovery and characterization of novel immunosuppressive ligands that facilitate immune evasion are crucial for developing more potent anti-cancer therapies.
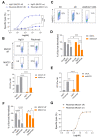
Methods: We conducted gain-of-function screens using a CRISPRa (CRISPR activation) library that covered the entire human transmembrane sub-genome to identify surface molecules capable of hindering NK-mediated cytotoxicity. The immunosuppressive role and mechanism of MUC21 were validated using NK and T cell mediated cytotoxicity assays. Bioinformatics tools were employed to assess the clinical implications of mucin-21 (MUC21) in cancer cell immunity.
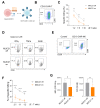
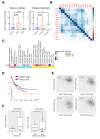
Results: Our genetic screens revealed that MUC21 expression on cancer cell surfaces inhibits both the cytotoxic activity of NK cells and antibody-dependent cellular cytotoxicity, but not affecting complement-dependent cytotoxicity. Additionally, MUC21 expression hinders T cell activation by impeding antigen recognition, thereby diminishing the effectiveness of the immune checkpoint inhibitor, anti-PD-L1. Moreover, MUC21 expression suppress the antitumor function of both CAR-T cells and CAR-NK cells. Mechanistically, MUC21 facilitates immune evasion by creating steric hindrance, preventing interactions between cancer and immune cells. Bioinformatics analysis revealed elevated MUC21 expression in lung cancer, which correlated with reduced infiltration and activation of cytotoxic immune cells. Intriguingly, MUC21 expression was higher in non-small cell lung cancer (NSCLC) tumors that were non-responsive to anti-PD-(L)1 treatment compared to responsive tumors.
Conclusions: These findings indicate that surface MUC21 serves as a potent immunosuppressive ligand, shielding cancer cells from NK and CD8+T cell attacks. This suggests that inhibiting MUC21 could be a promising strategy to improve cancer immunotherapy.
Keywords: CAR; CRISPR library screening; Cancer immunotherapy; MUC21.
© 2023. Italian National Cancer Institute ‘Regina Elena’.
Conflict of interest statement
All authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

