High-throughput chemogenetic drug screening reveals PKC-RhoA/PKN as a targetable signaling vulnerability in GNAQ-driven uveal melanoma
- PMID: 37858338
- PMCID: PMC10694608
- DOI: 10.1016/j.xcrm.2023.101244
High-throughput chemogenetic drug screening reveals PKC-RhoA/PKN as a targetable signaling vulnerability in GNAQ-driven uveal melanoma
Abstract
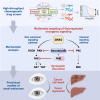
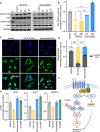
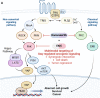
Uveal melanoma (UM) is the most prevalent cancer of the eye in adults, driven by activating mutation of GNAQ/GNA11; however, there are limited therapies against UM and metastatic UM (mUM). Here, we perform a high-throughput chemogenetic drug screen in GNAQ-mutant UM contrasted with BRAF-mutant cutaneous melanoma, defining the druggable landscape of these distinct melanoma subtypes. Across all compounds, darovasertib demonstrates the highest preferential activity against UM. Our investigation reveals that darovasertib potently inhibits PKC as well as PKN/PRK, an AGC kinase family that is part of the "dark kinome." We find that downstream of the Gαq-RhoA signaling axis, PKN converges with ROCK to control FAK, a mediator of non-canonical Gαq-driven signaling. Strikingly, darovasertib synergizes with FAK inhibitors to halt UM growth and promote cytotoxic cell death in vitro and in preclinical metastatic mouse models, thus exposing a signaling vulnerability that can be exploited as a multimodal precision therapy against mUM.
Keywords: FAK; GNAQ; PKC; PKN/PRK; chemogenetic drug screening; combination therapy; melanoma; precision medicine; synthetic lethality.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.S.G. reports consulting fees from Domain Pharmaceuticals, Pangea Therapeutics, and io9 and is founder of Kadima Pharmaceuticals, all unrelated to the current study. J.S.G. and N.A. hold patent US11679113B2 related in part to this work. The Krogan Laboratory has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. N.J.K. has financially compensated consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics, Interline Therapeutics, Rezo Therapeutics, Gen1E Lifesciences, Inc., and Twist Bioscience Corp. He is on the Board of Directors of Rezo Therapeutics and is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, and Interline Therapeutics. D.L.S. has a consulting agreement with Maze Therapeutics. J.B. is a consultant for Rocket Pharma. J.A.P. and S.C. are employees of Verastem, which has not influenced this study. Other authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

