Neuroimaging-based classification of PTSD using data-driven computational approaches: A multisite big data study from the ENIGMA-PGC PTSD consortium
- PMID: 37858907
- PMCID: PMC10842116
- DOI: 10.1016/j.neuroimage.2023.120412
Neuroimaging-based classification of PTSD using data-driven computational approaches: A multisite big data study from the ENIGMA-PGC PTSD consortium
Abstract
Background: Recent advances in data-driven computational approaches have been helpful in devising tools to objectively diagnose psychiatric disorders. However, current machine learning studies limited to small homogeneous samples, different methodologies, and different imaging collection protocols, limit the ability to directly compare and generalize their results. Here we aimed to classify individuals with PTSD versus controls and assess the generalizability using a large heterogeneous brain datasets from the ENIGMA-PGC PTSD Working group.
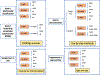
Methods: We analyzed brain MRI data from 3,477 structural-MRI; 2,495 resting state-fMRI; and 1,952 diffusion-MRI. First, we identified the brain features that best distinguish individuals with PTSD from controls using traditional machine learning methods. Second, we assessed the utility of the denoising variational autoencoder (DVAE) and evaluated its classification performance. Third, we assessed the generalizability and reproducibility of both models using leave-one-site-out cross-validation procedure for each modality.
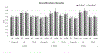
Results: We found lower performance in classifying PTSD vs. controls with data from over 20 sites (60 % test AUC for s-MRI, 59 % for rs-fMRI and 56 % for d-MRI), as compared to other studies run on single-site data. The performance increased when classifying PTSD from HC without trauma history in each modality (75 % AUC). The classification performance remained intact when applying the DVAE framework, which reduced the number of features. Finally, we found that the DVAE framework achieved better generalization to unseen datasets compared with the traditional machine learning frameworks, albeit performance was slightly above chance.
Conclusion: These results have the potential to provide a baseline classification performance for PTSD when using large scale neuroimaging datasets. Our findings show that the control group used can heavily affect classification performance. The DVAE framework provided better generalizability for the multi-site data. This may be more significant in clinical practice since the neuroimaging-based diagnostic DVAE classification models are much less site-specific, rendering them more generalizable.
Keywords: Classification; Deep learning; Machine learning; Multimodal MRI; Posttraumatic stress disorder.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest Dr. Thompson received partial grant support from Biogen, Inc., and Amazon, Inc., for work unrelated to the current study; Dr. Lebois reports unpaid membership on the Scientific Committee for International Society for the Study of Trauma and Dissociation (ISSTD), grant support from the National Institute of Mental Health, K01 MH118467 and the Julia Kasparian Fund for Neuroscience Research, McLean Hospital. Dr. Lebois also reports spousal IP payments from Vanderbilt University for technology licensed to Acadia Pharmaceuticals unrelated to the present work. ISSTD and NIMH were not involved in the analysis or preparation of the manuscript; Dr. Etkin reports salary and equity from Alto Neuroscience, equity from Mindstrong Health and Akili Interactive. Other authors have no conflicts of interest to declare.
Figures



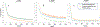


References
-
- Altmann A, Tolosi L, Sander O, et al., 2010. Permutation importance: a corrected feature importance measure. Bioinformatics 26, 1340–1347. - PubMed
-
- Bai X, Wang X, Liu X, et al., 2021. Explainable deep learning for efficient and robust pattern recognition: a survey of recent developments. Pattern Recognit. 120, 108102.
Publication types
MeSH terms
Grants and funding
- K01 MH122774/MH/NIMH NIH HHS/United States
- IK2 RX002922/RX/RRD VA/United States
- R33 MH108753/MH/NIMH NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- T32 GM007507/GM/NIGMS NIH HHS/United States
- R01 MH119227/MH/NIMH NIH HHS/United States
- F31 MH122047/MH/NIMH NIH HHS/United States
- T32 MH018931/MH/NIMH NIH HHS/United States
- R01 MH043454/MH/NIMH NIH HHS/United States
- P50 HD103556/HD/NICHD NIH HHS/United States
- K01 MH118467/MH/NIMH NIH HHS/United States
- R21 MH097784/MH/NIMH NIH HHS/United States
- R01 MH129832/MH/NIMH NIH HHS/United States
- R01 MH119132/MH/NIMH NIH HHS/United States
- R61 MH127005/MH/NIMH NIH HHS/United States
- R01 MH111671/MH/NIMH NIH HHS/United States
- U54 EB020403/EB/NIBIB NIH HHS/United States
- R01 MH117601/MH/NIMH NIH HHS/United States
- R01 AT011267/AT/NCCIH NIH HHS/United States
- K23 MH090366/MH/NIMH NIH HHS/United States
- R61 NS120249/NS/NINDS NIH HHS/United States
- IK2 CX001600/CX/CSRD VA/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

