MEK inhibitors in cancer treatment: structural insights, regulation, recent advances and future perspectives
- PMID: 37859720
- PMCID: PMC10583825
- DOI: 10.1039/d3md00145h
MEK inhibitors in cancer treatment: structural insights, regulation, recent advances and future perspectives
Abstract
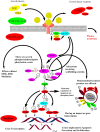
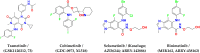
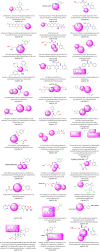
MEK1/2 are critical components of the RAS-RAF-MEK-ERK or MAPK signalling pathway that regulates a variety of cellular functions including proliferation, survival, and differentiation. In 1997, a lung cancer cell line was first found to have a MEK mutation (encoding MEK2P298L). MEK is involved in various human cancers such as non-small cell lung cancer (NSCLC), spurious melanoma, and pancreatic, colorectal, basal, breast, and liver cancer. To date, 4 MEK inhibitors i.e., trametinib, cobimetinib, selumetinib, and binimetinib have been approved by the FDA and several are under clinical trials. In this review, we have highlighted structural insights into the MEK1/2 proteins, such as the αC-helix, catalytic loop, P-loop, F-helix, hydrophobic pocket, and DFG motif. We have also discussed current issues with all FDA-approved MEK inhibitors or drugs under clinical trials and combination therapies to improve the efficacy of clinical drugs. Finally, this study addressed recent developments on synthetic MEK inhibitors (from their discovery in 1997 to 2022), their unique properties, and their relevance to MEK mutant inhibition.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials
Miscellaneous

