Identification of disease modules using higher-order network structure
- PMID: 37860106
- PMCID: PMC10582521
- DOI: 10.1093/bioadv/vbad140
Identification of disease modules using higher-order network structure
Abstract
Motivation: Higher-order interaction patterns among proteins have the potential to reveal mechanisms behind molecular processes and diseases. While clustering methods are used to identify functional groups within molecular interaction networks, these methods largely focus on edge density and do not explicitly take into consideration higher-order interactions. Disease genes in these networks have been shown to exhibit rich higher-order structure in their vicinity, and considering these higher-order interaction patterns in network clustering have the potential to reveal new disease-associated modules.
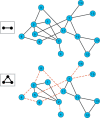
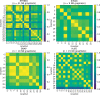
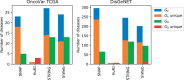
Results: We propose a higher-order community detection method which identifies community structure in networks with respect to specific higher-order connectivity patterns beyond edges. Higher-order community detection on four different protein-protein interaction networks identifies biologically significant modules and disease modules that conventional edge-based clustering methods fail to discover. Higher-order clusters also identify disease modules from genome-wide association study data, including new modules that were not discovered by top-performing approaches in a Disease Module DREAM Challenge. Our approach provides a more comprehensive view of community structure that enables us to predict new disease-gene associations.
Availability and implementation: https://github.com/Reed-CompBio/graphlet-clustering.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
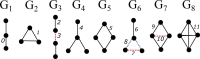



References
-
- Arenas A, Fernández A, Fortunato S. et al. Motif-based communities in complex networks. J Phys A Math Theor 2008;41:224001.
-
- Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995;57:289–300.
LinkOut - more resources
Full Text Sources

