Transcriptional profile reveals the physiological responses to prey availability in the mixotrophic chrysophyte Poterioochromonas malhamensis
- PMID: 37860135
- PMCID: PMC10582637
- DOI: 10.3389/fmicb.2023.1173541
Transcriptional profile reveals the physiological responses to prey availability in the mixotrophic chrysophyte Poterioochromonas malhamensis
Abstract
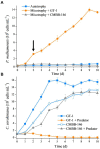
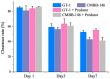
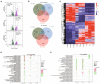
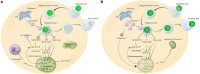
Mixotrophic flagellates, which have diverse nutritional modes and play important roles in connecting the microbial loop with the classical food chain, are ideal models to study the mechanisms of adaptation between different nutritional modes in protists. In their natural ecosystems, mixotrophic flagellates may encounter microalgal prey of different digestibility, which may affect the carbon flow. To date, a molecular biological view of the metabolic processes in the mixotrophic flagellate Poterioochromonas malhamensis during nutritional adaptation and feeding on microalgal prey of different digestibility is still lacking. Accordingly, this study focused on the gene expression differences in P. malhamensis under autotrophy, being fed by the digestible microalga Chlorella sorokiniana GT-1, and being fed by the indigestible microalga C. sorokiniana CMBB-146. Results showed that the growth rate of P. malhamensis under autotrophy was much lower than that when fed by digestible microalgae. Addition of C. sorokiniana CMBB-146 could only increase the growth rate of P. malhamensis in the first 3 days, but the cell concentration of P. malhamensis started to decrease gradually after 4 days. Compared to autotrophic P. malhamensis, total 6,583 and 3,510 genes were significantly and differentially expressed in P. malhamensis fed by digestible microalgae and indigestible microalgae, respectively. Compared to autotrophic cells, genes related to the ribosome, lysosome, glycolysis, gluconeogenesis, TCA cycle, β-oxidation, duplication, and β-1,3-glucan in P. malhamensis grazing on digestible prey were up-regulated, while genes related to light harvesting and key enzymes referring to chlorophyll were down-regulated. Genes related to apoptosis and necrosis in P. malhamensis were up-regulated after grazing on indigestible microalgae compared to the autotrophic group, which we suggest is associated with the up-regulation of genes related to lysosome enzymes. This study provides abundant information on the potential intracellular physiological responses of P. malhamensis during the process of nutritional adaptation.
Keywords: Poterioochromonas malhamensis; feeding behavior; physiology; prey availability; transcriptome.
Copyright © 2023 Ma, Yang, Chen, Ke, Gong and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data, Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom.
-
- Burkholder J. M., Glibert P. M., Skelton H. M. (2008). Mixotrophy, a major mode of nutrition for harmful algal species in eutrophic waters. Harmful Algae 8, 77–93. doi: 10.1016/j.hal.2008.08.010 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

