Identification and validation of PCSK9 as a prognostic and immune-related influencing factor in tumorigenesis: a pan-cancer analysis
- PMID: 37860186
- PMCID: PMC10584329
- DOI: 10.3389/fonc.2023.1134063
Identification and validation of PCSK9 as a prognostic and immune-related influencing factor in tumorigenesis: a pan-cancer analysis
Abstract
Introduction: Proprotein convertase subtilisin/kexin-9 (PCSK9) has been primarily studied in the cardiovascular field however, its role in cancer pathophysiology remains incompletely defined. Recently, a pivotal role for PCSK9 in cancer immunotherapy was proposed based on the finding that PCSK9 inhibition was associated with enhancing the antigen presentation efficacy of target programmed cell death-1 (PD-1). Herein, we provide results of a comprehensive pan-cancer analysis of PCSK9 that assessed its prognostic and immunological functions in cancer.
Methods: Using a variety of available online cancer-related databases including TIMER, cBioPortal, and GEPIA, we identified the abnormal expression of PCSK9 and its potential clinical associations in diverse cancer types including liver, brain and lung. We also validated its role in progression-free survival (PFS) and immune infiltration in neuroblastoma.
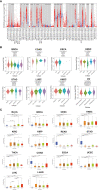
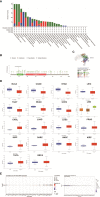
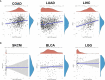
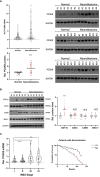
Results: Overall, the pan-cancer survival analysis revealed an association between dysregulated PCSK9 and poor clinical outcomes in various cancer types. Specifically, PCSK9 was extensively genetically altered across most cancer types and was consistently found in different tumor types and substages when compared with adjacent normal tissues. Thus, aberrant DNA methylation may be responsible for PCSK9 expression in many cancer types. Focusing on liver hepatocellular carcinoma (LIHC), we found that PCSK9 expression correlated with clinicopathological characteristics following stratified prognostic analyses. PCSK9 expression was significantly associated with immune infiltrate since specific markers of CD8+ T cells, macrophage polarization, and exhausted T cells exhibited different PCSK9-related immune infiltration patterns in LIHC and lung squamous cell carcinoma. In addition, PCSK9 was connected with resistance of drugs such as erlotinib and docetaxel. Finally, we validated PCSK9 expression in clinical neuroblastoma samples and concluded that PCSK9 appeared to correlate with a poor PFS and natural killer cell infiltration in neuroblastoma patients.
Conclusion: PCSK9 could serve as a robust prognostic pan-cancer biomarker given its correlation with immune infiltrates in different cancer types, thus potentially highlighting a new direction for targeted clinical therapy of cancers.
Keywords: immune infiltrate; pan-cancer; pcsk9; prognosis; tumorigenesis.
Copyright © 2023 Sun, Zhu, Shen, Huang, Li, Li, Ma and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

