Accelerating pharmaceutical R&D with a user-friendly AI system for histopathology image analysis
- PMID: 37860714
- PMCID: PMC10582575
- DOI: 10.1016/j.jpi.2023.100337
Accelerating pharmaceutical R&D with a user-friendly AI system for histopathology image analysis
Abstract
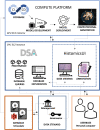
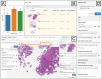
A system for analysis of histopathology data within a pharmaceutical R&D environment has been developed with the intention of enabling interdisciplinary collaboration. State-of-the-art AI tools have been deployed as easy-to-use self-service modules within an open-source whole slide image viewing platform, so that non-data scientist users (e.g., clinicians) can utilize and evaluate pre-trained algorithms and retrieve quantitative results. The outputs of analysis are automatically cataloged in the database to track data provenance and can be viewed interactively on the slide as annotations or heatmaps. Commonly used models for analysis of whole slide images including segmentation, extraction of hand-engineered features for segmented regions, and slide-level classification using multi-instance learning are included and new models can be added as needed. The source code that supports running inference with these models internally is backed up by a robust CI/CD pipeline to ensure model versioning, robust testing, and seamless deployment of the latest models. Examples of the use of this system in a pharmaceutical development workflow include glomeruli segmentation, enumeration of podocyte count from WT-1 immuno-histochemistry, measurement of beta-1 integrin target engagement from immunofluorescence, digital glomerular phenotyping from periodic acid-Schiff histology, PD-L1 score prediction using multi-instance learning, and the deployment of the open-source Segment Anything model to speed up annotation.
Keywords: Annotation; Model cataloging; Segment Anything; Segmentation; Visualization.
© 2023 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Wong E.T., et al. Outcomes and prognostic factors in recurrent glioma patients enrolled onto phase II clinical trials. J Clin Oncol. 1999;17(8):2572. - PubMed
-
- Granter S.R., Beck A.H., Papke D.J., Jr. AlphaGo, deep learning, and the future of the human microscopist. Arch Pathol Lab Med. 2017;141(5):619–621. - PubMed
-
- Henstock P.V. Artificial intelligence for pharma: time for internal investment. Trends Pharmacol Sci. 2019;40(8):543–546. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

