Tick-borne flavivirus NS5 antagonizes interferon signaling by inhibiting the catalytic activity of TYK2
- PMID: 37860832
- PMCID: PMC10702846
- DOI: 10.15252/embr.202357424
Tick-borne flavivirus NS5 antagonizes interferon signaling by inhibiting the catalytic activity of TYK2
Abstract
The mechanisms utilized by different flaviviruses to evade antiviral functions of interferons are varied and incompletely understood. Using virological approaches, biochemical assays, and mass spectrometry analyses, we report here that the NS5 protein of tick-borne encephalitis virus (TBEV) and Louping Ill virus (LIV), two related tick-borne flaviviruses, antagonize JAK-STAT signaling through interactions with the tyrosine kinase 2 (TYK2). Co-immunoprecipitation (co-IP) experiments, yeast gap-repair assays, computational protein-protein docking and functional studies identify a stretch of 10 residues of the RNA dependent RNA polymerase domain of tick-borne flavivirus NS5, but not mosquito-borne NS5, that is critical for interactions with the TYK2 kinase domain. Additional co-IP assays performed with several TYK2 orthologs reveal that the interaction is conserved across mammalian species. In vitro kinase assays show that TBEV and LIV NS5 reduce the catalytic activity of TYK2. Our results thus illustrate a novel mechanism by which viruses suppress the interferon response.
Keywords: JAK-STAT signaling; TYK2; emerging viruses; interferon evasion by viruses; tick-borne flaviviruses.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
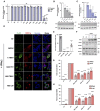
Huh7 cells were left uninfected (NI) or were infected with TBEV or LIV at a multiplicity of infection (MOI) of 0.5 and 0.05 respectively. Twenty‐four h later, the percentages of cells expressing the viral E protein were determined by flow cytometry analysis. Data are the means ± SD of three biological replicates. One‐way ANOVA tests were performed, ****P < 0.0001.
- B–D
Huh7 cells were left uninfected (NI) or infected for 24 h with TBEV or LIV, at a MOI of 0.5 and 0.05 respectively, and then treated or not with IFN⍺2 at 2,000 IU/ml for 8 h. The relative amounts of cell‐associated viral RNA (B) were measured by RT–qPCR analysis and were expressed as genome equivalents (GE) per μg of total cellular RNAs. The relative amounts of ISG15 (C) and ISG56 (D) mRNAs were determined by RT–qPCR analysis. Results were first normalized to GAPDH mRNA and then to non‐treated uninfected mRNA levels, which were set at 1. Data are the means ± SD of four biological replicates. One‐way ANOVA tests were performed, ns: non‐significant, ****P < 0.0001.
- E
Huh7 cells were left uninfected (NI) or were infected as in (A) for 24 h and stimulated or not with IFN⍺2 at 2,000 IU/ml for 30 min. Whole‐cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of three biological replicates.
- F
293T cells were left uninfected (NI) or were infected with TBEV or LIV at an MOI 0.02 for 48 h. They were stimulated or not with IFN⍺2 at 2,000 IU/ml for 30 min before fixation. They were stained with antibodies recognizing the viral E protein (green), STAT1p (red) and NucBlue® (blue). Images are representative of two biological replicates. Scale bars, 50 μm.
- A
293T cells were co‐transfected with the Firefly luciferase reporter plasmid p‐ISRE‐luc, TK Renilla luciferase control plasmid phRluc‐TK, and plasmids encoding individual viral proteins. Empty vectors (EV) and plasmids encoding the NS5 of YFV protein were used as negative and positive controls, respectively. Cells were stimulated 7 h post‐transfection with IFN⍺2 at 200 IU/ml and assayed for luciferase activity 16 h later. The data were analyzed by first normalizing the Firefly luciferase activity to the Renilla luciferase (Rluc) activity and then to EV samples, which were set at 100%. Data are means ± SD of three biological replicates. One‐way ANOVA tests with Dunnett's correction were performed, ns: non‐significant, ****P < 0.0001.
- B, C
293T cells were co‐transfected with Firefly luciferase reporter plasmid p‐ISRE‐luc, TK Renilla luciferase control plasmid phRluc‐TK and increasing amounts (ranging from 0.1 to 5 ng) of plasmids encoding the NS5 of TBEV (B) or LIV (C). Cells were stimulated 7 h post‐transfection with IFN⍺2 at 200 IU/ml and assayed for luciferase activity at 24 h post‐transfection. Cells transfected with empty vector (EV) were used as negative controls. The data were analyzed by first normalizing the Firefly luciferase activity to the Renilla luciferase (Rluc) activity and then to EV samples, which were set at 100%. Data are mean ± SD of three biological replicates. One‐way ANOVA tests with Dunnett's correction were performed, ns: non‐significant, **P < 0.01, ***P < 0.001, ****P < 0.0001. Western blot analyses were performed with anti‐FLAG and anti‐actin antibodies on the same samples. Non‐transfected (NT) cells added in the Western blotting analysis for comparison. Data are representative of at least three biological replicates.
- D, E
293T cells were mock‐transfected (NT), transfected with empty plasmid (EV) or plasmids expressing FLAG‐tagged NS5 from TBEV, LIV, or YFV. Cells transfected with plasmids expressing FLAG‐tagged C protein from LIV were used as negative controls. Twenty‐four hours later, they were left untreated (NT) or treated with IFN⍺2 at 2,000 IU/ml for 30 min before fixation. Cells were then stained with antibodies recognizing the FLAG tag of viral proteins (green), STAT1p (red) and NucBlue (blue). Images are representative of three biological replicates. Scale bars, 40 μm. (E) Percentages of STAT1p‐positive nuclei among cells expressing viral proteins were estimated by analyzing at least 15 fields (~150 cells) per condition. Data are mean ± SD. One‐way ANOVA tests with Dunnett's correction were performed. ****P < 0.0001.
- F
293T cells were mock‐transfected (NT), transfected with empty plasmid (EV) or plasmids expressing FLAG‐NS5 from TBEV, LIV or YFV for 24 h. Cells were left untreated (NT) or stimulated with IFN⍺2 (400 IU/ml) 30 min before harvest. Whole‐cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of three biological replicates.
- G, H
293T cells were mock‐transfected (NT), transfected with empty vectors (EV) or plasmids expressing FLAG‐NS5 from TBEV, LIV or YFV. Twenty‐four h later, they were left untreated (NT) or treated with IFN⍺2 (2,000 IU/ml) or IL29 (100 ng/μl) overnight. The relative amounts of ISG15 (G) and ISG56 (H) mRNAs were determined by RT–qPCR analysis. Results were first normalized to GAPDH mRNA and then to mRNA levels of treated cells transfected with EV, which were set at 100%. Data are means ± SD of three biological replicates. One‐way ANOVA tests with Dunnett's correction were performed. ****P < 0.0001.

Viral protein expression in 293T cells. 293T cells were mock‐transfected (NT), transfected with empty plasmids (EV) or with plasmids encoding FLAG‐tagged TBEV or LIV viral proteins. A plasmid coding a FLAG‐tagged version of YFV NS5 protein was included in the analysis. Cells were harvested 24 h post‐transfection and protein expression was assessed by Western blotting with anti‐FLAG and anti‐actin antibodies. Data are representative of three biological replicates. NS: nonstrucural proteins.
YFV NS5 protein reduces ISRE activation in 293T cells. 293T cells were co‐transfected with Firefly luciferase reporter plasmid p‐ISRE‐luc, TK Renilla luciferase control plasmid phRluc‐TK and increasing amounts (ranging from 0.1 to 70 ng) of plasmids encoding the NS5 of YFV. Cells were stimulated 7 h post‐transfection with IFN⍺2 at 200 IU/ml and assayed for luciferase activity at 24 h post‐transfection. Cells transfected with empty vector (EV) were used as negative controls. The data were analyzed by first normalizing the Firefly luciferase activity to the Renilla luciferase (Rluc) activity and then to EV samples, which were set at 100%. Data are mean ± SD of three biological replicates. One‐way ANOVA tests with Dunnett's correction were performed, ns: non‐significant ***P < 0.001, ****P < 0.0001. Western blot analyses were performed with anti‐FLAG and anti‐actin antibodies on the same samples. Non‐transfected (NT) cells added in the Western blotting analysis for comparison. Data are representative of at least three biological replicates.

293T cells were transfected with empty plasmids (EV) or with plasmids expressing FLAG‐tagged versions of TBEV or LIV NS5. Twenty‐four hours later, cell lysates were immunoprecipitated with anti‐FLAG magnetic beads. Immunoprecipitates were analyzed by Western blotting with antibodies against the FLAG tag. Data are representative of four biological replicates.
Mass spectrometry (MS) analysis was performed with four biological replicates per bait. Empty vector (EV) conditions were used as negative controls. Data were analyzed with three different MS scoring algorithms (see Results and Experimental procedures sections). Only proteins that were identified in 2 out of the 3 analyses were considered high confidence partners of NS5 and are represented here. Cellular partners common to TBEV and LIV NS5 are depicted in purple, while partners specific for TBEV or LIV NS5 are represented in darker purple and lighter purple, respectively. TYK2 is highlighted in yellow.
293T cells were infected for 24 h with TBEV or LIV at an MOI of 0.5 and 0.05 respectively. Cell lysates were immunoprecipitated using antibodies specific for NS5 of tick‐borne flaviviruses. Western blot analysis was performed on whole cells lysates (Input) and NS5‐immunoprecipitates (IP NS5) using the indicated antibodies. The presented western blot is representative of three biological replicates.

Huh7 cells were mock‐transfected (mock) or transfected with plasmids expressing FLAG‐tagged NS5 from TBEV or LIV. Twenty‐four hours later, cells were fixed and stained with antibodies recognizing the FLAG tag (green), and NucBlue® (blue). Images are representative of three biological replicates. Scale bars, 50 μm.
Huh7 cells were left uninfected (NI) or were infected with TBEV or LIV at an MOI 0.02 for 48 h. They were stimulated or not with IFN⍺2 at 2,000 IU/ml for 30 min before fixation. Cells were stained with antibodies recognizing the viral E protein (red), NS5 protein (green) and NucBlue® (blue). Images are representative of two biological replicates. Scale bars, 40 μm.

The flavivirus NS5 contains two domains: an N‐terminal methyltransferase (MTase) domain (30 kDa) and an RNA‐dependent RNA polymerase (RdRp) domain (90 kDa). The full‐length proteins and the domains were fused to an N‐terminal FLAG tag.
293T cells were mock‐transfected (NT), transfected with plasmids expressing either FLAG‐tagged full‐length (FL) NS5 or individual domains (MTase or RdRp), together with V5‐tagged TYK2 plasmid. Cells transfected with empty vectors (EV) or YFV NS5 plasmids were used as negative and positive controls, respectively. Cells were lysed 24 h post‐transfection. Western blot analyses were performed on whole‐cell lysates with the indicated antibodies (Input). Immunoprecipitation assays were performed on the same samples with anti‐FLAG magnetic beads (IP FLAG). Lysates were revealed using FLAG and V5 antibodies. Results are representative of three biological replicates.
The interactions between NS5 (full‐length or RdRp domains) and the C‐terminal domain of TYK2 were assessed by Yeast Gap Repair assays. Protein expression and interaction enable yeast growth on a medium deprived of leucine, tryptophan, and histidine and supplemented with 5 mM 3‐aminotriazole (3‐AT). Yeast transformed with circular pPC86 were used as positive recombination control (see Experimental procedures section) and yeasts transformed with linear pPC86 alone provided a negative control of interaction. Yeast expressing NS5 of Yellow Fever virus (FL YFV) was used as a negative interaction control. Results are representative of three biological replicates.
293T cells were co‐transfected with Firefly luciferase reporter plasmid p‐ISRE‐luc, TK Renilla luciferase control plasmid phRluc‐TK and plasmids expressing TBEV or LIV full‐length (FL) NS5 or individual domains (MTase or RdRp). Empty vectors (EV) and plasmids expressing YFV NS5 protein were used as negative and positive controls, respectively. Cells were stimulated 7 h post‐transfection with IFN⍺2 at 200 IU/ml and assayed for luciferase activity 24 h later. The data were analyzed by first normalizing the Firefly luciferase activity to the Renilla luciferase (Rluc) activity and then to EV samples, which were set at 100%. The data are means ± SD of three biological replicates. One‐way ANOVA tests with Dunnett's correction were performed. ns: non‐significant, **P < 0.01, ****P < 0.0001.
293T cells were mock‐transfected (NT), transfected with empty plasmids (EV) or plasmids expressing full‐length versions or individual domains of NS5 from TBEV or LIV for 24 h. Cells expressing YFV NS5 were used in parallel. Cells were left untreated (NT) or stimulated with IFN⍺2 (400 IU/ml) 30 min before lysis. Whole‐cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of three biological replicates.

293T cells were mock‐transfected (NT) or transfected with plasmids expressing FLAG‐tagged versions of NS5 from flaviviruses transmitted by ticks (TBEV, LIV and LGTV), Culex mosquitoes (WNV, USUV or JEV) or Aedes mosquitoes (ZIKV, DENV or YEV), together with empty plasmids (EV) or plasmids expressing TYK2‐V5. Cells were lysed 24 h post‐transfection. Western blot analysis was performed on whole‐cell lysates with the indicated antibodies (Input). Immunoprecipitation assays were performed on the same samples using anti‐FLAG magnetic beads (IP FLAG). Lysates were revealed using FLAG and V5 antibodies. Results are representative of three biological replicates.
Structure‐based alignment of tick‐ and mosquito‐borne flavivirus NS5, from amino acids 616 to 658. Red boxed residues in the variable region are the residues predicted to interact with TYK2 after molecular docking simulations.
Superimposed structures of the NS5 RdRp domain of YFV (PDB:6QSN; blue) and TBEV (PDB: 7D6N; green). NS5 YFV presents an insertion (DES) located within the variable region that extends the alpha helix with negatively charged surface‐exposed residues (dotted red circle).
Protein sequence of TBEV and YFV NS5 (from amino acids 616 to 658), as well as of a TBEV‐VRYFV construct where TBEV residues from the variable region (VR) within the inter B‐C domain have been replaced by the corresponding YFV aa.
293T cells were co‐transfected with Firefly luciferase reporter plasmid p‐ISRE‐luc, TK Renilla luciferase control plasmid phRluc‐TK and full‐length TBEV NS5 (wild‐type or TBEV‐VRYFV mutant) or empty vectors (EV) as negative controls. Cells were stimulated 7 h post‐transfection with IFN⍺2 at 200 IU/ml and assayed for luciferase activity at 24 h post‐transfection. The data were analyzed by first normalizing the Firefly luciferase activity to the Renilla luciferase (Rluc) activity and then to EV samples, which were set at 100%. Data are mean ± SD of three biological replicates. One‐way ANOVA tests with Dunnett's correction were performed, ns: non‐significant, ****P < 0.0001.
293T cells were mock‐transfected (NT), transfected with plasmids encoding FLAG‐tagged versions of NS5 TBEV (full‐length or RdRp domain), either wild‐type or TBEV VRYFV mutant, together with empty plasmid (EV) or plasmid encoding TYK2‐V5. Cells were lysed 24 h post‐transfection. Western blot analysis was performed on whole‐cell lysates with the indicated antibodies (Input). Immunoprecipitation assays were performed on the same samples with anti‐FLAG magnetic beads (IP FLAG). Lysates were revealed using FLAG and V5 antibodies. Results are representative of three biological replicates.
Huh7 cells were mocked‐electoporated or electoporated with in‐vitro synthetized RNAs derived from wild‐type (rTBEV) or mutated TBEV replicons (rTBEV‐VR‐YFV or rTBEV‐GAA). Three days later the relative amounts of cell‐associated viral RNA were measured by RT–qPCR analysis and were expressed as genome equivalents (GE) per μg of total cellular RNAs. Data are mean ± SD of three biological replicates. T‐tests were performed, ns: non‐significant, **P < 0.05.
Huh7 cells were mocked‐electoporated or electoporated with in‐vitro synthetized RNA derived from wild‐type or mutated TBEV replicons (VR‐YFV or GAA). Three days later, they were stimulated or not with IFN⍺2 (2,000 IU/ml) for 30 min. Whole‐cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of three biological replicates.


293T cells were co‐transfected or not (NT) with plasmids expressing FLAG‐tagged versions of NS5 TBEV or LIV together with V5‐tagged TYK2 from different mammalian species or control empty vectors. Cells were lysed 24 h post‐transfection. Western blot analysis was performed on cell lysates with the indicated antibodies (Input). Immunoprecipitation assays were performed on the same samples using anti‐FLAG magnetic beads (IP FLAG). Immunoprecipitates were revealed with anti‐FLAG and anti‐V5 antibodies. Results are representative of three biological replicates.
Interactions between NS5 from TBEV and LIV with the C‐terminal domain of human, bovine, sheep, and goat TYK2 were assessed by yeast Gap Repair assays. Protein expression and interaction enable yeast growth on a medium devoid of leucine, tryptophan, and histidine and supplemented with 5 mM 3‐aminotriazole (3‐AT). Yeast transformed with circular pPC86 were used as positive recombination control (see Experimental procedures section) and yeast transformed with linear pPC86 alone provided a negative control of interaction. Results are representative of three biological replicates.
TYK2 is composed of four domains: an N‐terminal FERM domain, a SH2 domain, a kinase‐like (KL) domain and a C‐terminal tyrosine kinase (TK) domain. Mutants of TYK2 were generated as indicated. All are carrying a C‐terminal V5 tag.
293T cells were mock‐transfected (NT), transfected with empty plasmids (EV) or plasmids expressing full‐length versions of TBEV or LIV NS5, together with different versions of V5‐tagged TYK2 plasmids. Cells were lysed 24 h post‐transfection. Western blot analysis was performed on whole‐cell lysates with the indicated antibodies (Input). Immunoprecipitation assays were performed on the same samples using anti‐FLAG magnetic beads (IP FLAG). Lysates were revealed using FLAG and V5 antibodies. Results are representative of three biological replicates.
Structural cartoon of the interaction between the TBEV RdRp domain (green) and the kinase domain of TYK2 (gray), as predicted by molecular docking. Data presented in Fig 5 suggest that TBEV NS5 RdRp interacts with the TYK2 via a variable region of its B‐C loop (yellow). Intermolecular hydrogen bonds are represented as dotted lines in the enlarged box. The residue K930, which is involved in ATP binding and kinase activity, is colored in magenta.
293T cells were mock‐transfected (NT), transfected with empty plasmids (EV) or plasmids expressing NS5 from TBEV, LIV or YFV. Twenty‐four hours later, cells were stimulated with IFN⍺2 (400 IU/ml) for 15 min. Whole‐cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. Data are representative of three biological replicates. For densitometric analysis of band intensities see Fig EV5A.
TYK2 kinase activity was assessed in vitro using the kinase domain (aa 871–1,187) of TYK2, a peptide derived from IRS‐1 as substrate and ATP. Kinase reactions were carried with or without recombinant NS5 proteins as indicated. The level of ATP, which decreases during the kinase reaction, was inversely correlated to the luciferase activity. The activity of TYK2 in the absence of viral proteins was set at 100%. The data are means ± SD of at least six biological replicates. One‐way ANOVA tests with Dunnett's correction were performed, ns: non‐significant, ****P < 0.0001.
In vitro kinase assays were performed as in (G) by replacing IRS‐1 peptides by recombinant STAT1. The samples were analyzed by Western blot with the indicated antibodies. Data are representative of three biological replicates. For densitometric analysis of band intensities see Fig EV5C.


Densitometric analysis of Western blots from three biological replicates showing the relative abundances of pTYK2 to total TYK2. Data are expressed as a percentage of the values of cells transfected with an empty plasmid (EV). They are means ± SD. One‐way ANOVA tests with Dunnett's correction were performed. ns: non‐significant, ***P < 0.001.
Viral protein expression in vitro. Coomassie Gel of purified NS5 (full‐length protein or individual domain).
Densitometric analysis of Western blots from three biological replicates showing the relative abundances of pSTAT1 to total STAT1. Data are expressed as a percentage of the samples containing no NS5 (mock), and are means ± SD. One‐way ANOVA tests with Dunnett's correction were performed, ns: non‐significant, **P < 0.01.
References
-
- Burfoot MS, Rogers NC, Watling D, Smith JM, Pons S, Paonessaw G, Pellegrini S, White MF, Kerr IM (1997) Janus kinase‐dependent activation of insulin receptor substrate 1 in response to interleukin‐4, oncostatin M, and the interferons. J Biol Chem 272: 24183–24190 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

