Genetic modifiers of repeat expansion disorders
- PMID: 37861103
- PMCID: PMC10754329
- DOI: 10.1042/ETLS20230015
Genetic modifiers of repeat expansion disorders
Abstract
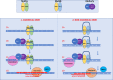
Repeat expansion disorders (REDs) are monogenic diseases caused by a sequence of repetitive DNA expanding above a pathogenic threshold. A common feature of the REDs is a strong genotype-phenotype correlation in which a major determinant of age at onset (AAO) and disease progression is the length of the inherited repeat tract. Over a disease-gene carrier's life, the length of the repeat can expand in somatic cells, through the process of somatic expansion which is hypothesised to drive disease progression. Despite being monogenic, individual REDs are phenotypically variable, and exploring what genetic modifying factors drive this phenotypic variability has illuminated key pathogenic mechanisms that are common to this group of diseases. Disease phenotypes are affected by the cognate gene in which the expansion is found, the location of the repeat sequence in coding or non-coding regions and by the presence of repeat sequence interruptions. Human genetic data, mouse models and in vitro models have implicated the disease-modifying effect of DNA repair pathways via the mechanisms of somatic mutation of the repeat tract. As such, developing an understanding of these pathways in the context of expanded repeats could lead to future disease-modifying therapies for REDs.
Keywords: DNA synthesis and repair; cag repeat; genetic modifier; repeat expansion; somatic DNA expansion; somatic instability.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures
References
-
- Scahill, R.I., Zeun, P., Osborne-Crowley, K., Johnson, E.B., Gregory, S., Parker, C.et al. (2020) Biological and clinical characteristics of gene carriers far from predicted onset in the Huntington's disease young adult study (HD-YAS): a cross-sectional analysis. Lancet Neurol. 19, 502–512 10.1016/S1474-4422(20)30143-5 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
