Plasma cell-free DNA methylation analysis for ovarian cancer detection: Analysis of samples from a case-control study and an ovarian cancer screening trial
- PMID: 37861205
- PMCID: PMC7617350
- DOI: 10.1002/ijc.34757
Plasma cell-free DNA methylation analysis for ovarian cancer detection: Analysis of samples from a case-control study and an ovarian cancer screening trial
Abstract
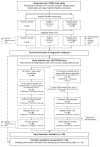
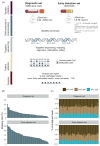
Analysis of cell-free DNA methylation (cfDNAme), alone or combined with CA125, could help to detect ovarian cancers earlier and may reduce mortality. We assessed cfDNAme in regions of ZNF154, C2CD4D and WNT6 via targeted bisulfite sequencing in diagnostic and early detection (preceding diagnosis) settings. Diagnostic samples were obtained via prospective blood collection in cell-free DNA tubes in a convenience series of patients with a pelvic mass. Early detection samples were matched case-control samples derived from the UK Familial Ovarian Cancer Screening Study (UKFOCSS). In the diagnostic set (ncases = 27, ncontrols = 41), the specificity of cfDNAme was 97.6% (95% CI: 87.1%-99.9%). High-risk cancers were detected with a sensitivity of 80% (56.3%-94.3%). Combination of cfDNAme and CA125 resulted in a sensitivity of 94.4% (72.7%-99.9%) for high-risk cancers. Despite technical issues in the early detection set (ncases = 29, ncontrols = 29), the specificity of cfDNAme was 100% (88.1%-100.0%). We detected 27.3% (6.0%-61.0%) of high-risk cases with relatively lower genomic DNA (gDNA) contamination. The sensitivity rose to 33.3% (7.5%-70.1%) in samples taken <1 year before diagnosis. We detected ovarian cancer in several patients up to 1 year before diagnosis despite technical limitations associated with archival samples (UKFOCSS). Combined cfDNAme and CA125 assessment may improve ovarian cancer screening in high-risk populations, but future large-scale prospective studies will be required to validate current findings.
Keywords: cell-free DNA; diagnosis; methylation; ovarian cancer.
© 2023 The Authors. International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Conflict of interest statement
Martin Widschwendter, Allison Jones, Iona Evans and Tobias Paprotka are named as inventors on a patent which describes the DNAme markers analysed in this manuscript. Andreas Leimbach and Markus Schmitt are employed by Eurofins Genomics, which has submitted patent applications related to the intellectual property outlined in this manuscript. Kristina Gemzell-Danielsson has served on an ad hoc basis as speaker or expert on meetings organised by Exelgyn/Nordic, Concept Foundation and HRA-Pharma. Aleksandra Gentry-Maharaj has been funded by grants from the Medical Research Council, Cancer Research UK, National Institute for Health Research and The Eve Appeal; she reports funded research collaborations with industry—iLOF (intelligent Lab on Fibre), RNA Guardian, Micronoma, Mercy Bioanalytics and academics—Cambridge University, QIMR Berghofer Medical Research Institute, Imperial College London, University of Innsbruck and Dana Farber, USA. Aleksandra Gentry-Maharaj is a member of ACED Gynaecological Cancer Working Group and is ACED Co-Director Research Domain Trials. Sophia Apostolidou has been funded by grants from the Medical Research Council, Cancer Research UK, National Institute for Health Research and National Institute for Health Research Health Technology Assessment and has been funded by Abcodia until July 2021 (paid to UCL); she reports funded research collaborations with Cambridge University, QIMR Berghofer Medical Research Institute, iLOF (intelligent Lab on Fibre), RNA Guardian, Micronoma, Mercy Bioanalytics, Syntent Biotechnology, Imperial College London, Dana Faber Cancer Institute and National Health and Medical Research Council Australia. Usha Menon reports institutional research collaborations in early detection of ovarian cancer with industry—RNA Guardian, Micronoma, Mercy Bioanalytics, Synteny and research collaborations in early detection of cancer, in particular ovarian cancer with UK, US and Australian academics supported by public and charity funded grants.
The other authors declare no conflict of interest.
Figures


References
-
- Ovarian cancer survival statistics. 2022. [Internet] https://www.cancerresearchuk.org/health-professional/cancer-statistics/s....
-
- Menon U, Ryan A, Kalsi J, et al. Risk algorithm using serial biomarker measurements doubles the number of screen-detected cancers compared with a single-threshold rule in the United Kingdom collaborative trial of ovarian cancer screening. J Clin Oncol. 2015;33:2062–2071. doi: 10.1200/JCO.2014.59.4945. - DOI - PMC - PubMed
-
- Menon U, Gentry-Maharaj A, Burnell M, et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK collaborative trial of ovarian cancer screening (UKCTOCS): a randomised controlled trial. Lancet. 2021;397:2182–2193. doi: 10.1016/S0140-6736(21)00731-5. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

