Computational analysis of crosstalk between transcriptional regulators and RNA-binding proteins suggests mutual regulation of polycomb proteins and SRSF1 influencing adult hippocampal neurogenesis
- PMID: 37861946
- PMCID: PMC10501017
- DOI: 10.1007/s44192-023-00034-5
Computational analysis of crosstalk between transcriptional regulators and RNA-binding proteins suggests mutual regulation of polycomb proteins and SRSF1 influencing adult hippocampal neurogenesis
Abstract
Background: Adult hippocampal neurogenesis (AHN) is a clinically significant neural phenomenon. Understanding its molecular regulation would be important. In this regard, most studies have focused on transcriptional regulators (TRs), epigenetic modifiers, or non-coding RNAs. RNA-binding proteins (RBPs) have emerged as dominant molecular regulators. It would be significant to understand the potential cross-talk between RBPs and TRs, which could influence AHN.
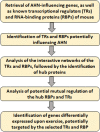
Methods: The present study employed computational analyses to identify RBPs and TRs regulating AHN, followed by the analysis of their interaction networks and detection of hub proteins. Next, the potential mutual regulation of hub TRs and RBPs was analyzed. Additionally, hippocampal genes differentially expressed upon exercise were analyzed for potential regulation by the identified TRs and RBPs.
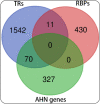
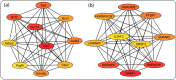
Results: 105 TRs and 26 RBPs were found to influence AHN, which could also form interactive networks. Polycomb complex proteins were among the TR network hubs, while HNRNP and SRSF family members were among the hub RBPs. Further, the polycomb complex proteins and SRSF1 could have a mutual regulatory relationship, suggesting a cross-talk between epigenetic/transcriptional and post-transcriptional regulatory pathways. A number of exercise-induced hippocampal genes were also found to be potential targets of the identified TRs and RBPs.
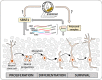
Conclusion: SRSF1 may influence post-transcriptional stability, localization, and alternative splicing patterns of polycomb complex transcripts, and the polycomb proteins may in turn epigenetically influence the SRSF1. Further experimental validation of these regulatory loops/networks could provide novel insights into the molecular regulation of AHN, and unravel new targets for disease-treatment.
Keywords: Adult hippocampal neurogenesis (AHN); Physical exercise; Polycomb complex; Post-transcriptional regulation; RNA-binding proteins (RBPs); SRSF.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources

