Protein interaction network revealed by quantitative proteomic analysis links TFIIB to multiple aspects of the transcription cycle
- PMID: 37863410
- PMCID: PMC10872477
- DOI: 10.1016/j.bbapap.2023.140968
Protein interaction network revealed by quantitative proteomic analysis links TFIIB to multiple aspects of the transcription cycle
Abstract
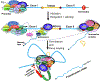
Although TFIIB is widely regarded as an initiation factor, recent reports have implicated it in multiple aspects of eukaryotic transcription. To investigate the broader role of TFIIB in transcription, we performed quantitative proteomic analysis of yeast TFIIB. We purified two different populations of TFIIB; one from soluble cell lysate, which is not engaged in transcription, and the other from the chromatin fraction which yields the transcriptionally active form of the protein. TFIIB purified from the chromatin exhibits several interactions that explain its non-canonical roles in transcription. RNAPII, TFIIF and TFIIH were the only components of the preinitiation complex with a significant presence in chromatin TFIIB. A notable feature was enrichment of all subunits of CF1 and Rat1 3' end processing-termination complexes in chromatin-TFIIB preparation. Subunits of the CPF termination complex were also detected in both chromatin and soluble derived TFIIB preparations. These results may explain the presence of TFIIB at the 3' end of genes during transcription as well as its role in promoter-termination interaction.
Keywords: Budding yeast; Proteomic analyses; RNA polymerase II; TFIIB; Transcription.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflicts of interests.
Figures

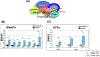
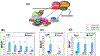
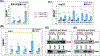
Similar articles
-
TFIIB-Termination Factor Interaction Affects Termination of Transcription on Genome-Wide Scale.Int J Mol Sci. 2024 Aug 8;25(16):8643. doi: 10.3390/ijms25168643. Int J Mol Sci. 2024. PMID: 39201330 Free PMC article.
-
Evidence for a complex of transcription factor IIB with poly(A) polymerase and cleavage factor 1 subunits required for gene looping.J Biol Chem. 2011 Sep 30;286(39):33709-18. doi: 10.1074/jbc.M110.193870. Epub 2011 Aug 11. J Biol Chem. 2011. PMID: 21835917 Free PMC article.
-
Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF.Mol Cell Biol. 2012 Jan;32(1):12-25. doi: 10.1128/MCB.06242-11. Epub 2011 Oct 24. Mol Cell Biol. 2012. PMID: 22025674 Free PMC article.
-
Beyond the canonical role of TFIIB in eukaryotic transcription.Curr Genet. 2022 Feb;68(1):61-67. doi: 10.1007/s00294-021-01223-x. Epub 2021 Nov 19. Curr Genet. 2022. PMID: 34797379 Free PMC article. Review.
-
Rethinking the role of TFIIF in transcript initiation by RNA polymerase II.Transcription. 2012 Jul-Aug;3(4):156-9. doi: 10.4161/trns.20725. Epub 2012 Jul 1. Transcription. 2012. PMID: 22771986 Free PMC article. Review.
Cited by
-
The Central FacilitaTOR: Coordinating Transcription and Translation in Eukaryotes.Int J Mol Sci. 2025 Mar 21;26(7):2845. doi: 10.3390/ijms26072845. Int J Mol Sci. 2025. PMID: 40243440 Free PMC article. Review.
-
Genome-wide analysis of TFIIB's role in termination of transcription.Res Sq [Preprint]. 2024 Jul 15:rs.3.rs-4619136. doi: 10.21203/rs.3.rs-4619136/v1. Res Sq. 2024. PMID: 39070618 Free PMC article. Preprint.
-
TFIIB-Termination Factor Interaction Affects Termination of Transcription on Genome-Wide Scale.Int J Mol Sci. 2024 Aug 8;25(16):8643. doi: 10.3390/ijms25168643. Int J Mol Sci. 2024. PMID: 39201330 Free PMC article.
-
idopNetwork Analysis of Salt-Responsive Transcriptomes Reveals Hub Regulatory Modules and Genes in Populus euphratica.Int J Mol Sci. 2025 Apr 25;26(9):4091. doi: 10.3390/ijms26094091. Int J Mol Sci. 2025. PMID: 40362331 Free PMC article.
-
Sub2: A TFIIB Interacting Protein with Pleiotropic Roles in mRNA Synthesis, Processing and Transport.Int J Biochem Physiol. 2024;9(1):250. doi: 10.23880/ijbp-16000250. Epub 2024 Jun 6. Int J Biochem Physiol. 2024. PMID: 40330028 Free PMC article.
References
-
- Al-Husini N, Medler S, Ansari A, Crosstalk of promoter and terminator during RNA polymerase II transcription cycle, Biochim. Biophys. Acta (BBA)-Gene Regulat. Mechan. 194657 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

