Proteomic Serum Profiling of Holstein Friesian Cows with Different Pathological Forms of Bovine Paratuberculosis Reveals Changes in the Acute-Phase Response and Lipid Metabolism
- PMID: 37863471
- PMCID: PMC11301775
- DOI: 10.1021/acs.jproteome.3c00244
Proteomic Serum Profiling of Holstein Friesian Cows with Different Pathological Forms of Bovine Paratuberculosis Reveals Changes in the Acute-Phase Response and Lipid Metabolism
Abstract
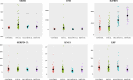
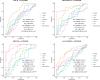
The lack of sensitive diagnostic methods to detect Mycobacterium avium subsp. paratuberculosis (Map) subclinical infections has hindered the control of paratuberculosis (PTB). The serum proteomic profiles of naturally infected cows presenting focal and diffuse pathological forms of PTB and negative controls (n = 4 per group) were analyzed using TMT-6plex quantitative proteomics. Focal and diffuse are the most frequent pathological forms in subclinical and clinical stages of PTB, respectively. One (focal versus (vs.) control), eight (diffuse vs. control), and four (focal vs. diffuse) differentially abundant (DA) proteins (q-value < 0.05) were identified. Ingenuity pathway analysis of the DA proteins revealed changes in the acute-phase response and lipid metabolism. Six candidate biomarkers were selected for further validation by specific ELISA using serum from animals with focal, multifocal, and diffuse PTB-associated lesions (n = 108) and controls (n = 56). Overall, the trends of the serum expression levels of the selected proteins were consistent with the proteomic results. Alpha-1-acid glycoprotein (ORM1)-based ELISA, insulin-like growth factor-binding protein 2 (IGFBP2)-based ELISA, and the anti-Map ELISA had the best diagnostic performance for detection of animals with focal, multifocal, and diffuse lesions, respectively. Our findings identify potential biomarkers that improve diagnostic sensitivity of PTB and help to elucidate the mechanisms involved in PTB pathogenesis.
Keywords: Mycobacterium avium subsp. paratuberculosis; acute-phase proteins; biomarkers; cattle; diagnosis; disease progression; paratuberculosis; pathogenesis; serum proteome.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
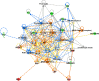
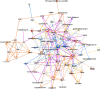
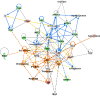
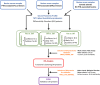
References
-
- Tiwari A.; VanLeeuwen J. A.; Dohoo I. R.; Stryhn H.; Keefe G. P.; Haddad J. P. Effects of seropositivity for bovine leukemia virus, bovine viral diarrhoea virus, Mycobacterium avium subspecies paratuberculosis, and Neospora caninum on culling in dairy cattle in four Canadian provinces. Vet. Microbiol. 2005, 109 (3–4), 147–58. 10.1016/j.vetmic.2005.05.011. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

