Control of TGFβ signalling by ubiquitination independent function of E3 ubiquitin ligase TRIP12
- PMID: 37863914
- PMCID: PMC10589240
- DOI: 10.1038/s41419-023-06215-y
Control of TGFβ signalling by ubiquitination independent function of E3 ubiquitin ligase TRIP12
Abstract
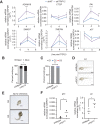
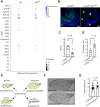
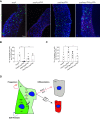
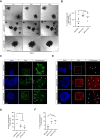
Transforming growth factor β (TGFβ) pathway is a master regulator of cell proliferation, differentiation, and death. Deregulation of TGFβ signalling is well established in several human diseases including autoimmune disorders and cancer. Thus, understanding molecular pathways governing TGFβ signalling may help better understand the underlying causes of some of those conditions. Here, we show that a HECT domain E3 ubiquitin ligase TRIP12 controls TGFβ signalling in multiple models. Interestingly, TRIP12 control of TGFβ signalling is completely independent of its E3 ubiquitin ligase activity. Instead, TRIP12 recruits SMURF2 to SMAD4, which is most likely responsible for inhibitory monoubiquitination of SMAD4, since SMAD4 monoubiquitination and its interaction with SMURF2 were dramatically downregulated in TRIP12-/- cells. Additionally, genetic inhibition of TRIP12 in human and murine cells leads to robust activation of TGFβ signalling which was rescued by re-introducing wildtype TRIP12 or a catalytically inactive C1959A mutant. Importantly, TRIP12 control of TGFβ signalling is evolutionary conserved. Indeed, genetic inhibition of Drosophila TRIP12 orthologue, ctrip, in gut leads to a reduced number of intestinal stem cells which was compensated by the increase in differentiated enteroendocrine cells. These effects were completely normalised in Drosophila strain where ctrip was co-inhibited together with Drosophila SMAD4 orthologue, Medea. Similarly, in murine 3D intestinal organoids, CRISPR/Cas9 mediated genetic targeting of Trip12 enhances TGFβ mediated proliferation arrest and cell death. Finally, CRISPR/Cas9 mediated genetic targeting of TRIP12 in MDA-MB-231 breast cancer cells enhances the TGFβ induced migratory capacity of these cells which was rescued to the wildtype level by re-introducing wildtype TRIP12. Our work establishes TRIP12 as an evolutionary conserved modulator of TGFβ signalling in health and disease.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

