The transcription factor Zeb1 controls homeostasis and function of type 1 conventional dendritic cells
- PMID: 37863917
- PMCID: PMC10589231
- DOI: 10.1038/s41467-023-42428-7
The transcription factor Zeb1 controls homeostasis and function of type 1 conventional dendritic cells
Abstract
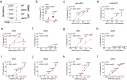
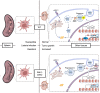
Type 1 conventional dendritic cells (cDC1) are the most efficient cross-presenting cells that induce protective cytotoxic T cell response. However, the regulation of their homeostasis and function is incompletely understood. Here we observe a selective reduction of splenic cDC1 accompanied by excessive cell death in mice with Zeb1 deficiency in dendritic cells, rendering the mice more resistant to Listeria infection. Additionally, cDC1 from other sources of Zeb1-deficient mice display impaired cross-presentation of exogenous antigens, compromising antitumor CD8+ T cell responses. Mechanistically, Zeb1 represses the expression of microRNA-96/182 that target Cybb mRNA of NADPH oxidase Nox2, and consequently facilitates reactive-oxygen-species-dependent rupture of phagosomal membrane to allow antigen export to the cytosol. Cybb re-expression in Zeb1-deficient cDC1 fully restores the defective cross-presentation while microRNA-96/182 overexpression in Zeb1-sufficient cDC1 inhibits cross-presentation. Therefore, our results identify a Zeb1-microRNA-96/182-Cybb pathway that controls cross-presentation in cDC1 and uncover an essential role of Zeb1 in cDC1 homeostasis.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

