ERRα promotes glycolytic metabolism and targets the NLRP3/caspase-1/GSDMD pathway to regulate pyroptosis in endometrial cancer
- PMID: 37864196
- PMCID: PMC10588109
- DOI: 10.1186/s13046-023-02834-7
ERRα promotes glycolytic metabolism and targets the NLRP3/caspase-1/GSDMD pathway to regulate pyroptosis in endometrial cancer
Erratum in
-
Correction: ERRα promotes glycolytic metabolism and targets the NLRP3/caspase-1/GSDMD pathway to regulate pyroptosis in endometrial cancer.J Exp Clin Cancer Res. 2025 Jan 25;44(1):23. doi: 10.1186/s13046-025-03292-z. J Exp Clin Cancer Res. 2025. PMID: 39856770 Free PMC article. No abstract available.
Abstract
Background: Tumor cells can resist chemotherapy-induced pyroptosis through glycolytic reprogramming. Estrogen-related receptor alpha (ERRα) is a central regulator of cellular energy metabolism associated with poor cancer prognosis. Herein, we refine the oncogenic role of ERRα in the pyroptosis pathway and glycolytic metabolism.
Methods: The interaction between ERRα and HIF-1α was verified using co-immunoprecipitation. The transcriptional binding sites of ERRα and NLRP3 were confirmed using dual-luciferase reporter assay and cleavage under targets and tagmentation (CUT&Tag). Flow cytometry, transmission electron microscopy, scanning electron microscopy, cell mito stress test, and extracellular acidification rate analysis were performed to investigate the effects of ERRα on the pyroptosis pathway and glycolytic metabolism. The results of these experiments were further confirmed in endometrial cancer (EC)-derived organoids and nude mice. In addition, the expression of ERRα-related pyroptosis genes was analyzed using The Cancer Genome Atlas and Gene Expression Omnibus database.
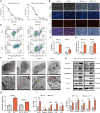
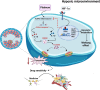
Results: Triggered by a hypoxic microenvironment, highly expressed ERRα could bind to the promoter of NLRP3 and inhibit caspase-1/GSDMD signaling, which reduced inflammasome activation and increased pyroptosis resistance, thereby resulting in the resistance of cancer cells to cisplatin. Moreover, ERRα activated glycolytic rate-limiting enzyme to bridge glycolytic metabolism and pyroptosis in EC. This phenomenon was further confirmed in EC-derived organoids and nude mice. CUT & Tag sequencing and The Cancer Genome Atlas database analysis showed that ERRα participated in glycolysis and programmed cell death, which resulted in EC progression.
Conclusions: ERRα inhibits pyroptosis in an NLRP3-dependent manner and induces glycolytic metabolism, resulting in cisplatin resistance in EC cells.
Keywords: Cisplatin resistance; ERRα; Endometrial cancer; Metabolic reprogramming; Pyroptosis.
© 2023. Italian National Cancer Institute ‘Regina Elena’.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures





References
-
- Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression[J]. Int J Radiat Biol. 2019;95(7):912–9. - PubMed
-
- Sun L, Suo C, Li S, et al. Metabolic reprogramming for cancer cells and their microenvironment: beyond the Warburg effect[J]. Biochim Biophys Acta Rev Cancer. 2018;1870(1):51–66. - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFC2704303/Fund of National Key R&D Program of China
- Grant no. 82203739/National Nature Science Foundation of China
- grant no. 2021ZQNZD011/Fujian Health Young and Middle-aged Scientific Research Major Project
- Grant no. 2020J02059/the Natural Science Foundation of Fujian Province
- 2021J01404/the Natural Science Foundation of Fujian Province
LinkOut - more resources
Full Text Sources

