Lysine 117 on ataxin-3 modulates toxicity in Drosophila models of Spinocerebellar Ataxia Type 3
- PMID: 37865002
- PMCID: PMC10841544
- DOI: 10.1016/j.jns.2023.120828
Lysine 117 on ataxin-3 modulates toxicity in Drosophila models of Spinocerebellar Ataxia Type 3
Abstract
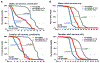
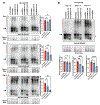
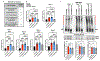
Ataxin-3 (Atxn3) is a deubiquitinase with a polyglutamine (polyQ) repeat tract whose abnormal expansion causes the neurodegenerative disease, Spinocerebellar Ataxia Type 3 (SCA3; also known as Machado-Joseph Disease). The ubiquitin chain cleavage properties of Atxn3 are enhanced when the enzyme is itself ubiquitinated at lysine (K) at position 117: in vitro, K117-ubiqutinated Atxn3 cleaves poly-ubiquitin markedly more rapidly compared to its unmodified counterpart. How polyQ expansion causes SCA3 remains unclear. To gather insights into the biology of disease of SCA3, here we posited the question: is K117 important for toxicity caused by pathogenic Atxn3? To answer this question, we generated transgenic Drosophila lines that express full-length, human, pathogenic Atxn3 with 80 polyQ with an intact or mutated K117. We found that mutating K117 mildly enhances the toxicity and aggregation of pathogenic Atxn3. An additional transgenic line that expresses Atxn3 without any K residues confirms increased aggregation of pathogenic Atxn3 whose ubiquitination is perturbed. These findings suggest that Atxn3 ubiquitination is a regulatory step of SCA3, in part by modulating its aggregation.
Keywords: Aggregation; Ataxia; Deubiquitinase; Polyglutamine; Ubiquitin.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they do not have any conflicts of interest to disclose.
Figures




Update of
-
Lysine 117 on ataxin-3 modulates toxicity in Drosophila models of Spinocerebellar Ataxia Type 3.bioRxiv [Preprint]. 2023 Jun 1:2023.05.30.542896. doi: 10.1101/2023.05.30.542896. bioRxiv. 2023. Update in: J Neurol Sci. 2023 Nov 15;454:120828. doi: 10.1016/j.jns.2023.120828. PMID: 37398109 Free PMC article. Updated. Preprint.
References
-
- Brand AH, et al. , 1994. Ectopic expression in Drosophila. Methods Cell Biol. 44, 635–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

