Using CRISPR-Cas9/phosphoproteomics to identify substrates of calcium/calmodulin-dependent kinase 2δ
- PMID: 37865316
- PMCID: PMC10783575
- DOI: 10.1016/j.jbc.2023.105371
Using CRISPR-Cas9/phosphoproteomics to identify substrates of calcium/calmodulin-dependent kinase 2δ
Abstract
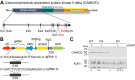
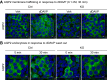
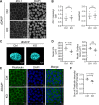
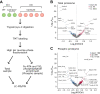
Ca2+/Calmodulin-dependent protein kinase 2 (CAMK2) family proteins are involved in the regulation of cellular processes in a variety of tissues including brain, heart, liver, and kidney. One member, CAMK2δ (CAMK2D), has been proposed to be involved in vasopressin signaling in the renal collecting duct, which controls water excretion through regulation of the water channel aquaporin-2 (AQP2). To identify CAMK2D target proteins in renal collecting duct cells (mpkCCD), we deleted Camk2d and carried out LC-MS/MS-based quantitative phosphoproteomics. Specifically, we used CRISPR/Cas9 with two different guide RNAs targeting the CAMK2D catalytic domain to create multiple CAMK2D KO cell lines. AQP2 protein abundance was lower in the CAMK2D KO cells than in CAMK2D-intact controls. AQP2 phosphorylation at Ser256 and Ser269 (normalized for total AQP2) was decreased. However, trafficking of AQP2 to and from the apical plasma membrane was sustained. Large-scale quantitative phosphoproteomic analysis (TMT-labeling) in the presence of the vasopressin analog dDAVP (0.1 nM, 30 min) allowed quantification of 11,570 phosphosites of which 169 were significantly decreased, while 206 were increased in abundance in CAMK2D KO clones. These data are available for browsing or download at https://esbl.nhlbi.nih.gov/Databases/CAMK2D-proteome/. Motif analysis of the decreased phosphorylation sites revealed a target preference of -(R/K)-X-X-p(S/T)-X-(D/E), matching the motif identified in previous in vitro phosphorylation studies using recombinant CAMK2D. Thirty five of the significantly downregulated phosphorylation sites in CAMK2D KO cells had exactly this motif and are judged to be likely direct CAMK2D targets. This adds to the list of known CAMK2D target proteins found in prior reductionist studies.
Keywords: aquaporin-2; mass spectrometry; vasopressin.
Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Soderling T.R., Chang B., Brickey D. Cellular signaling through multifunctional Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 2001;276:3719–3722. - PubMed
-
- Braun A.P., Schulman H. The multifunctional calcium/calmodulin-dependent protein kinase: from form to function. Annu. Rev. Physiol. 1995;57:417–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

