The heparin-binding hemagglutinin protein of Mycobacterium tuberculosis is a nucleoid-associated protein
- PMID: 37865319
- PMCID: PMC10665949
- DOI: 10.1016/j.jbc.2023.105364
The heparin-binding hemagglutinin protein of Mycobacterium tuberculosis is a nucleoid-associated protein
Abstract
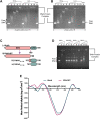
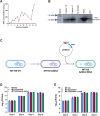
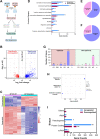
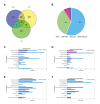
Nucleoid-associated proteins (NAPs) regulate multiple cellular processes such as gene expression, virulence, and dormancy throughout bacterial species. NAPs help in the survival and adaptation of Mycobacterium tuberculosis (Mtb) within the host. Fourteen NAPs have been identified in Escherichia coli; however, only seven NAPs are documented in Mtb. Given its complex lifestyle, it is reasonable to assume that Mtb would encode for more NAPs. Using bioinformatics tools and biochemical experiments, we have identified the heparin-binding hemagglutinin (HbhA) protein of Mtb as a novel sequence-independent DNA-binding protein which has previously been characterized as an adhesion molecule required for extrapulmonary dissemination. Deleting the carboxy-terminal domain of HbhA resulted in a complete loss of its DNA-binding activity. Atomic force microscopy showed HbhA-mediated architectural modulations in the DNA, which may play a regulatory role in transcription and genome organization. Our results showed that HbhA colocalizes with the nucleoid region of Mtb. Transcriptomics analyses of a hbhA KO strain revealed that it regulates the expression of ∼36% of total and ∼29% of essential genes. Deletion of hbhA resulted in the upregulation of ∼73% of all differentially expressed genes, belonging to multiple pathways suggesting it to be a global repressor. The results show that HbhA is a nonessential NAP regulating gene expression globally and acting as a plausible transcriptional repressor.
Keywords: DNA damage; DNA topology; DNA transcription; DNA-protein interaction; Mycobacterium tuberculosis; genome organization; nucleoid-associated proteins (NAPs).
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Luijsterburg M.S., Noom M.C., Wuite G.J., Dame R.T. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 2006;156:262–272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

