SHaploseek is a sequencing-only, high-resolution method for comprehensive preimplantation genetic testing
- PMID: 37865712
- PMCID: PMC10590366
- DOI: 10.1038/s41598-023-45292-z
SHaploseek is a sequencing-only, high-resolution method for comprehensive preimplantation genetic testing
Abstract
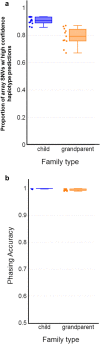
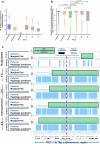
Recent advances in genomic technologies expand the scope and efficiency of preimplantation genetic testing (PGT). We previously developed Haploseek, a clinically-validated, variant-agnostic comprehensive PGT solution. Haploseek is based on microarray genotyping of the embryo's parents and relatives, combined with low-pass sequencing of the embryos. Here, to increase throughput and versatility, we aimed to develop a sequencing-only implementation of Haploseek. Accordingly, we developed SHaploseek, a universal PGT method to determine genome-wide haplotypes of each embryo based on low-pass (≤ 5x) sequencing of the parents and relative(s) along with ultra-low-pass (0.2-0.4x) sequencing of the embryos. We used SHaploseek to analyze five single lymphoblast cells and 31 embryos. We validated the genome-wide haplotype predictions against either bulk DNA, Haploseek, or, at focal genomic sites, PCR-based PGT results. SHaploseek achieved > 99% concordance with bulk DNA in two families from which single cells were derived from grown-up children. In embryos from 12 PGT families, all of SHaploseek's focal site haplotype predictions were concordant with clinical PCR-based PGT results. Genome-wide, there was > 99% median concordance between Haploseek and SHaploseek's haplotype predictions. Concordance remained high at all assayed sequencing depths ≥ 2x, as well as with only 1ng of parental DNA input. In subtelomeric regions, significantly more haplotype predictions were high-confidence in SHaploseek compared to Haploseek. In summary, SHaploseek constitutes a single-platform, accurate, and cost-effective comprehensive PGT solution.
© 2023. Springer Nature Limited.
Conflict of interest statement
D.B. is an employee and shareholder at The Janssen Pharmaceutical Companies of Johnson & Johnson. All other authors report no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

