Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease
- PMID: 37866628
- PMCID: PMC10692723
- DOI: 10.1016/j.jbc.2023.105382
Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease
Abstract
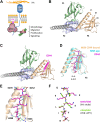
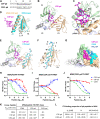
Proteomic studies have identified moesin (MSN), a protein containing a four-point-one, ezrin, radixin, moesin (FERM) domain, and the receptor CD44 as hub proteins found within a coexpression module strongly linked to Alzheimer's disease (AD) traits and microglia. These proteins are more abundant in Alzheimer's patient brains, and their levels are positively correlated with cognitive decline, amyloid plaque deposition, and neurofibrillary tangle burden. The MSN FERM domain interacts with the phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2) and the cytoplasmic tail of CD44. Inhibiting the MSN-CD44 interaction may help limit AD-associated neuronal damage. Here, we investigated the feasibility of developing inhibitors that target this protein-protein interaction. We have employed structural, mutational, and phage-display studies to examine how CD44 binds to the FERM domain of MSN. Interestingly, we have identified an allosteric site located close to the PIP2 binding pocket that influences CD44 binding. These findings suggest a mechanism in which PIP2 binding to the FERM domain stimulates CD44 binding through an allosteric effect, leading to the formation of a neighboring pocket capable of accommodating a receptor tail. Furthermore, high-throughput screening of a chemical library identified two compounds that disrupt the MSN-CD44 interaction. One compound series was further optimized for biochemical activity, specificity, and solubility. Our results suggest that the FERM domain holds potential as a drug development target. Small molecule preliminary leads generated from this study could serve as a foundation for additional medicinal chemistry efforts with the goal of controlling microglial activity in AD by modifying the MSN-CD44 interaction.
Keywords: Alzheimer disease; CD44; FERM domain; MSN; crystal structure; fluorescence resonance energy transfer; high-throughput screening; inhibitor; microglia; moesin; structure-activity relationship.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



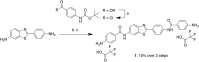
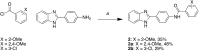
Update of
-
Development of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic strategy for Alzheimer's disease.bioRxiv [Preprint]. 2023 May 22:2023.05.22.541727. doi: 10.1101/2023.05.22.541727. bioRxiv. 2023. Update in: J Biol Chem. 2023 Dec;299(12):105382. doi: 10.1016/j.jbc.2023.105382. PMID: 37292860 Free PMC article. Updated. Preprint.
References
-
- Patterson C. The State of the Art of Dementia Research: New Frontiers. An Analysis of Prevalence, Incidence, Cost and Trends. Alzheimer’s Disease International (ADI); London: 2018.
-
- The Lancet Lecanemab for Alzheimer's disease: tempering hype and hope. Lancet. 2022;400:1899. - PubMed
-
- van Dyck C.H., Sabbagh M., Cohen S. Lecanemab in early Alzheimer's disease. N. Engl. J. Med. 2023;388:1631–1632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

