Transcription Factor Id1 Plays an Essential Role in Th9 Cell Differentiation by Inhibiting Tcf3 and Tcf4
- PMID: 37867222
- PMCID: PMC10724384
- DOI: 10.1002/advs.202305527
Transcription Factor Id1 Plays an Essential Role in Th9 Cell Differentiation by Inhibiting Tcf3 and Tcf4
Abstract
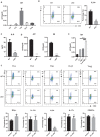
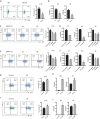
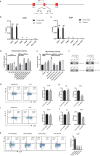
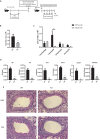
T helper type 9 (Th9) cells play important roles in immune responses by producing interleukin-9 (IL-9). Several transcription factors are responsible for Th9 cell differentiation; however, transcriptional regulation of Th9 cells is not fully understood. Here, it is shown that Id1 is an essential transcriptional regulator of Th9 cell differentiation. Id1 is induced by IL-4 and TGF-β. Id1-deficient naïve CD4 T cells fail to differentiate into Th9 cells, and overexpression of Id1 induce expression of IL-9. Mass spectrometry analysis reveals that Id1 interacts with Tcf3 and Tcf4 in Th9 cells. In addition, RNA-sequencing, chromatin immunoprecipitation, and transient reporter assay reveal that Tcf3 and Tcf4 bind to the promoter region of the Il9 gene to suppress its expression, and that Id1 inhibits their function, leading to Th9 differentiation. Finally, Id1-deficient Th9 cells ameliorate airway inflammation in an animal model of asthma. Thus, Id1 is a transcription factor that plays an essential role in Th9 cell differentiation by inhibiting Tcf3 and Tcf4.
Keywords: Id1; Tcf3; Tcf4; Th9 cell; differentiation; transcriptional regulation.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- a) Licona‐Limón P., Henao‐Mejia J., Temann A. U., Gagliani N., Licona‐Limón I., Ishigame H., Hao L., Herbert D. R., Flavell R. A., Immunity 2013, 39, 744 ; - PMC - PubMed
- b) Kaplan M. H., Hufford M. M., Olson M. R., Nat. Rev. Immunol. 2015, 15, 295 ; - PMC - PubMed
- c) Schmitt E., Bopp T., Semin Immunopathol 2017, 39, 5. - PubMed
-
- a) Staudt V., Bothur E., Klein M., Lingnau K., Reuter S., Grebe N., Gerlitzki B., Hoffmann M., Ulges A., Taube C., Dehzad N., Becker M., Stassen M., Steinborn A., Lohoff M., Schild H., Schmitt E., Bopp T., Immunity 2010, 33, 192 ; - PubMed
- b) Jones C. P., Gregory L. G., Causton B., Campbell G. A., Lloyd C. M., J. Allergy Clin. Immunol. 2012, 129, 1000. - PMC - PubMed
-
- a) Lu Y., Hong S., Li H., Park J., Hong B., Wang L., Zheng Y., Liu Z., Xu J., He J., Yang J., Qian J., Yi Q., J. Clin. Invest. 2012, 122, 4160; - PMC - PubMed
- b) Purwar R., Schlapbach C., Xiao S., Kang H. S., Elyaman W., Jiang X., Jetten A. M., Khoury S. J., Fuhlbrigge R. C., Kuchroo V. K., Clark R. A., Kupper T. S., Nat. Med. 2012, 18, 1248. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
