Cryo-electron microscopy in the fight against COVID-19-mechanism of virus entry
- PMID: 37867557
- PMCID: PMC10587472
- DOI: 10.3389/fmolb.2023.1252529
Cryo-electron microscopy in the fight against COVID-19-mechanism of virus entry
Abstract
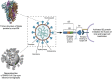
Cryogenic electron microscopy (cryo-EM) and electron tomography (cryo-ET) have become a critical tool for studying viral particles. Cryo-EM has enhanced our understanding of viral assembly and replication processes at a molecular resolution. Meanwhile, in situ cryo-ET has been used to investigate how viruses attach to and invade host cells. These advances have significantly contributed to our knowledge of viral biology. Particularly, prompt elucidations of structures of the SARS-CoV-2 spike protein and its variants have directly impacted the development of vaccines and therapeutic measures. This review discusses the progress made by cryo-EM based technologies in comprehending the severe acute respiratory syndrome coronavirus-2 (SARS-Cov-2), the virus responsible for the devastating global COVID-19 pandemic in 2020 with focus on the SARS-CoV-2 spike protein and the mechanisms of the virus entry and replication.
Keywords: ACE2; COVID-19; SARS-CoV-2; cryo-EM; cryo-ET; spike protein.
Copyright © 2023 Bodakuntla, Kuhn, Biertümpfel and Mizuno.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

