Allosteric crosstalk in modular proteins: Function fine-tuning and drug design
- PMID: 37867971
- PMCID: PMC10589753
- DOI: 10.1016/j.csbj.2023.10.013
Allosteric crosstalk in modular proteins: Function fine-tuning and drug design
Abstract
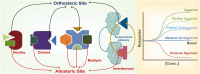
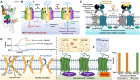
Modular proteins are regulatory proteins that carry out more than one function. These proteins upregulate or downregulate a biochemical cascade to establish homeostasis in cells. To switch the function or alter the efficiency (based on cellular needs), these proteins require different facilitators that bind to a site different from the catalytic (active/orthosteric) site, aka 'allosteric site', and fine-tune their function. These facilitators (or effectors) are allosteric modulators. In this Review, we have discussed the allostery, characterized them based on their mechanisms, and discussed how allostery plays an important role in the activity modulation and function fine-tuning of proteins. Recently there is an emergence in the discovery of allosteric drugs. We have also emphasized the role, significance, and future of allostery in therapeutic applications.
Keywords: Allostery; Drug design; Enzyme kinetics; Multi-domain proteins; Protein function regulation; Therapeutics.
© 2023 The Authors.
Conflict of interest statement
Authors declare no competing interest.
Figures




References
-
- Niks D., Hilario E., Dierkers A., Ngo H., Borchardt D., Neubauer T.J., et al. Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states. Biochemistry. 2013;52:6396–6411. doi: 10.1021/bi400795e. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
