Refined control of CRISPR-Cas9 gene editing in Clostridium sporogenes: the creation of recombinant strains for therapeutic applications
- PMID: 37869009
- PMCID: PMC10585264
- DOI: 10.3389/fimmu.2023.1241632
Refined control of CRISPR-Cas9 gene editing in Clostridium sporogenes: the creation of recombinant strains for therapeutic applications
Abstract
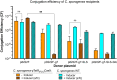
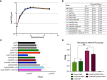
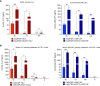
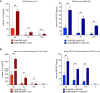
Despite considerable clinical success, the potential of cancer immunotherapy is restricted by a lack of tumour-targeting strategies. Treatment requires systemic delivery of cytokines or antibodies at high levels to achieve clinically effective doses at malignant sites. This is exacerbated by poor penetration of tumour tissue by therapeutic antibodies. High-grade immune-related adverse events (irAEs) occur in a significant number of patients (5-15%, cancer- and therapeutic-dependent) that can lead to lifelong issues and can exclude from treatment patients with pre-existing autoimmune diseases. Tumour-homing bacteria, genetically engineered to produce therapeutics, is one of the approaches that seeks to mitigate these drawbacks. The ability of Clostridium sporogenes to form spores that are unable to germinate in the presence of oxygen (typical of healthy tissue) offers a unique advantage over other vectors. However, the limited utility of existing gene editing tools hinders the development of therapeutic strains. To overcome the limitations of previous systems, expression of the Cas9 protein and the gRNA was controlled using tetracycline inducible promoters. Furthermore, the components of the system were divided across two plasmids, improving the efficiency of cloning and conjugation. Genome integrated therapeutic genes were assayed biochemically and in cell-based functional assays. The potency of these strains was further improved through rationally-conceived gene knock-outs. The new system was validated by demonstrating the efficient addition and deletion of large sequences from the genome. This included the creation of recombinant strains expressing two pro-inflammatory cytokines, interleukin-2 (IL-2) and granulocyte macrophage-colony stimulating factor (GM-CSF), and a pro-drug converting enzyme (PCE). A comparative, temporal in vitro analysis of the integrant strains and their plasmid-based equivalents revealed a substantial reduction of cytokine activity in chromosome-based constructs. To compensate for this loss, a 7.6 kb operon of proteolytic genes was deleted from the genome. The resultant knock-out strains showed an 8- to 10-fold increase in cytokine activity compared to parental strains.
Keywords: CRISPR-Cas9; Clostridium sporogenes; cancer; cytokines; immunotherapy; secretion.
Copyright © 2023 Kubiak, Claessen, Zhang, Khazaie and Bailey.
Conflict of interest statement
AK was an employee of Exomnis Biotech B.V. at the time of the study design and execution. AK is an inventor on two patents pending WO2021123391 and WO2022171904. TB is an inventor on one patent pending WO2022171904. LC was an employee of Exomnis Biotech B.V. at the time of the study design and execution 2021/2022. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

