Antimicrobial activity, membrane interaction and structural features of short arginine-rich antimicrobial peptides
- PMID: 37869668
- PMCID: PMC10585156
- DOI: 10.3389/fmicb.2023.1244325
Antimicrobial activity, membrane interaction and structural features of short arginine-rich antimicrobial peptides
Abstract
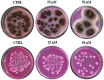
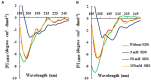
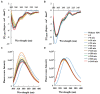
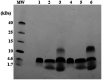
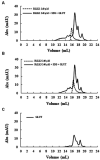
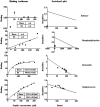
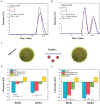
Antimicrobial activity of many AMPs can be improved by lysine-to-arginine substitution due to a more favourable interaction of arginine guanidinium moiety with bacterial membranes. In a previous work, the structural and functional characterization of an amphipathic antimicrobial peptide named RiLK1, including lysine and arginine as the positively charged amino acids in its sequence, was reported. Specifically, RiLK1 retained its β-sheet structure under a wide range of environmental conditions (temperature, pH, and ionic strength), and exhibited bactericidal activity against Gram-positive and Gram-negative bacteria and fungal pathogens with no evidence of toxicity on mammalian cells. To further elucidate the influence of a lysine-to-arginine replacement on RiLK1 conformational properties, antimicrobial activity and peptide-liposome interaction, a new RiLK1-derivative, named RiLK3, in which the lysine is replaced with an arginine residue, was projected and characterised in comparison with its parental compound. The results evidenced that lysine-to-arginine mutation not only did not assure an improvement in the antimicrobial potency of RiLK1 in terms of bactericidal, virucidal and fungicidal activities, but rather it was completely abolished against the hepatitis A virus. Therefore, RiLK1 exhibited a wide range of antimicrobial activity like other cationic peptides, although the exact mechanisms of action are not completely understood. Moreover, tryptophan fluorescence measurements confirmed that RiLK3 bound to negatively charged lipid vesicles with an affinity lower than that of RiLK1, although no substantial differences from the structural and self-assembled point of view were evidenced. Therefore, our findings imply that antimicrobial efficacy and selectivity are affected by several complex and interrelated factors related to substitution of lysine with arginine, such as their relative proportion and position. In this context, this study could provide a better rationalisation for the optimization of antimicrobial peptide sequences, paving the way for the development of novel AMPs with broad applications.
Keywords: antimicrobial compound; arginine; cationic arginine-rich peptide; membrane interaction; spectroscopy.
Copyright © 2023 Agrillo, Porritiello, Gratino, Balestrieri, Proroga, Mancusi, Cozzi, Vicenza, Dardano, Miranda, Escribá, Gogliettino and Palmieri.
Conflict of interest statement
BA was employed by Ampure S.R.L. PE is a founder of Laminar Pharmaceuticals. GP is a scientific consultant of Materias S.R.L. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Functional interplay between short antimicrobial peptides and model lipid membranes.Bioorg Chem. 2024 Dec;153:107939. doi: 10.1016/j.bioorg.2024.107939. Epub 2024 Nov 3. Bioorg Chem. 2024. PMID: 39520786
-
Effects of lysine-to-arginine substitution on antimicrobial activity of cationic stapled heptapeptides.Arch Pharm Res. 2018 Nov;41(11):1092-1097. doi: 10.1007/s12272-018-1084-5. Epub 2018 Oct 25. Arch Pharm Res. 2018. PMID: 30361948
-
Effect of Spacer Length Modification of the Cationic Side Chain on the Energetics of Antimicrobial Peptide Binding to Membrane-Mimetic Bilayers.J Chem Inf Model. 2023 Sep 25;63(18):5823-5833. doi: 10.1021/acs.jcim.3c01080. Epub 2023 Sep 8. J Chem Inf Model. 2023. PMID: 37684221
-
Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action.Biochim Biophys Acta. 2006 Sep;1758(9):1184-202. doi: 10.1016/j.bbamem.2006.04.006. Epub 2006 Apr 21. Biochim Biophys Acta. 2006. PMID: 16756942 Review.
-
Towards a structure-function analysis of bovine lactoferricin and related tryptophan- and arginine-containing peptides.Biochem Cell Biol. 2002;80(1):49-63. doi: 10.1139/o01-213. Biochem Cell Biol. 2002. PMID: 11908643 Review.
Cited by
-
A Reliable Multifaceted Solution against Foodborne Viral Infections: The Case of RiLK1 Decapeptide.Molecules. 2024 May 14;29(10):2305. doi: 10.3390/molecules29102305. Molecules. 2024. PMID: 38792166 Free PMC article.
-
Discovery of a Potent Antimicrobial Peptide Through Rational Design: A New Frontier in Pathogen Control.Biomolecules. 2025 Jul 11;15(7):989. doi: 10.3390/biom15070989. Biomolecules. 2025. PMID: 40723861 Free PMC article.
-
Unlocking the power of membrane biophysics: enhancing the study of antimicrobial peptides activity and selectivity.Biophys Rev. 2025 Apr 12;17(2):605-625. doi: 10.1007/s12551-025-01312-y. eCollection 2025 Apr. Biophys Rev. 2025. PMID: 40376398 Free PMC article. Review.
-
Engineering of antimicrobial peptide Brevinin-1pl: arginine, lysine, and histidine substitutions enhance antimicrobial-anticancer efficacy with reduced cytotoxicity.Front Chem. 2025 May 19;13:1579097. doi: 10.3389/fchem.2025.1579097. eCollection 2025. Front Chem. 2025. PMID: 40458657 Free PMC article.
-
Senegalin-2: A Novel Hexadecapeptide from Kassina senegalensis with Antibacterial and Muscle Relaxant Activities, and Its Derivative Senegalin-2BK as a Bradykinin Antagonist.Biomolecules. 2024 Dec 30;15(1):30. doi: 10.3390/biom15010030. Biomolecules. 2024. PMID: 39858425 Free PMC article.
References
-
- Agrillo B., Proroga Y. T. R., Gogliettino M., Balestrieri M., Tatè R., Nicolais L., et al. . (2020). A safe and multitasking antimicrobial decapeptide: the road from de novo design to structural and functional characterization. Int. J. Mol. Sci. 21:6952. doi: 10.3390/ijms21186952, PMID: - DOI - PMC - PubMed
-
- Ambrosio R. L., Rosselló C. A., Casares D., Palmieri G., Anastasio A., Escribá P. V. (2022). The antimicrobial peptide 1018-K6 interacts distinctly with eukaryotic and bacterial membranes, the basis of its specificity and bactericidal activity. Int. J. Mol. Sci. 23:12392. doi: 10.3390/ijms232012392, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

