An outlook to sophisticated technologies and novel developments for metabolic regulation in the Saccharomyces cerevisiae expression system
- PMID: 37869712
- PMCID: PMC10586203
- DOI: 10.3389/fbioe.2023.1249841
An outlook to sophisticated technologies and novel developments for metabolic regulation in the Saccharomyces cerevisiae expression system
Abstract
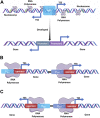
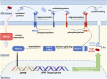
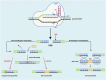
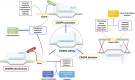
Saccharomyces cerevisiae is one of the most extensively used biosynthetic systems for the production of diverse bioproducts, especially biotherapeutics and recombinant proteins. Because the expression and insertion of foreign genes are always impaired by the endogenous factors of Saccharomyces cerevisiae and nonproductive procedures, various technologies have been developed to enhance the strength and efficiency of transcription and facilitate gene editing procedures. Thus, the limitations that block heterologous protein secretion have been overcome. Highly efficient promoters responsible for the initiation of transcription and the accurate regulation of expression have been developed that can be precisely regulated with synthetic promoters and double promoter expression systems. Appropriate codon optimization and harmonization for adaption to the genomic codon abundance of S. cerevisiae are expected to further improve the transcription and translation efficiency. Efficient and accurate translocation can be achieved by fusing a specifically designed signal peptide to an upstream foreign gene to facilitate the secretion of newly synthesized proteins. In addition to the widely applied promoter engineering technology and the clear mechanism of the endoplasmic reticulum secretory pathway, the innovative genome editing technique CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated system) and its derivative tools allow for more precise and efficient gene disruption, site-directed mutation, and foreign gene insertion. This review focuses on sophisticated engineering techniques and emerging genetic technologies developed for the accurate metabolic regulation of the S. cerevisiae expression system.
Keywords: CRISPR/Cas9; codon optimization; fusion partner; genomic engineering; metabolic regulation; promoter engineering.
Copyright © 2023 Wu, Feng, Sun, Hu, Jia and Zeng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Easy efficient HDR-based targeted knock-in in Saccharomyces cerevisiae genome using CRISPR-Cas9 system.Bioengineered. 2022 Jun;13(6):14857-14871. doi: 10.1080/21655979.2022.2162667. Bioengineered. 2022. PMID: 36602175 Free PMC article.
-
Genome Editing in Clostridium saccharoperbutylacetonicum N1-4 with the CRISPR-Cas9 System.Appl Environ Microbiol. 2017 May 1;83(10):e00233-17. doi: 10.1128/AEM.00233-17. Print 2017 May 15. Appl Environ Microbiol. 2017. PMID: 28258147 Free PMC article.
-
CRISPR system in the yeast Saccharomyces cerevisiae and its application in the bioproduction of useful chemicals.World J Microbiol Biotechnol. 2019 Jul 6;35(7):111. doi: 10.1007/s11274-019-2688-8. World J Microbiol Biotechnol. 2019. PMID: 31280424 Review.
-
A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae.Metab Eng. 2016 Jan;33:19-27. doi: 10.1016/j.ymben.2015.10.011. Epub 2015 Nov 4. Metab Eng. 2016. PMID: 26546089
-
Recent advances in metabolic engineering of Saccharomyces cerevisiae: New tools and their applications.Metab Eng. 2018 Nov;50:85-108. doi: 10.1016/j.ymben.2018.04.011. Epub 2018 Apr 25. Metab Eng. 2018. PMID: 29702275 Review.
Cited by
-
Recombinant protein production in Pseudoalteromonas haloplanktis TAC125 biofilm.Biofilm. 2024 Jan 24;7:100179. doi: 10.1016/j.bioflm.2024.100179. eCollection 2024 Jun. Biofilm. 2024. PMID: 38322580 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

