Highly-Specific Single-Stranded Oligonucleotides and Functional Nanoprobes for Clinical Determination of Chlamydia Trachomatis and Neisseria Gonorrhoeae Infections
- PMID: 37870167
- PMCID: PMC10754082
- DOI: 10.1002/advs.202304009
Highly-Specific Single-Stranded Oligonucleotides and Functional Nanoprobes for Clinical Determination of Chlamydia Trachomatis and Neisseria Gonorrhoeae Infections
Abstract
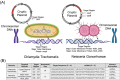
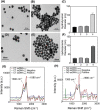
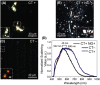
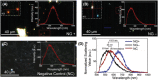
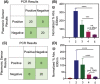
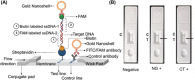
Early detection of Chlamydia trachomatis (CT) and Neisseria gonorrhoeae (NG) is the key to controlling the spread of these bacterial infections. An important step in developing biosensors involves identifying reliable sensing probes against specific genetic targets for CT and NG. Here, the authors have designed single-stranded oligonucleotides (ssDNAs) targeting mutually conserved genetic regions of cryptic plasmid and chromosomal DNA of both CT and NG. The 5'- and 3'- ends of these ssDNAs are differentially functionalized with thiol groups and coupled with gold nanoparticles (AuNP) to develop absorbance-based assay. The AuNPs agglomerate selectively in the presence of its target DNA sequence and demonstrate a change in their surface plasmon resonance. The optimized assay is then used to detect both CT and NG DNA extracted from 60 anonymized clinical samples with a clinical sensitivity of ∼100%. The limit of detection of the assays are found to be 7 and 5 copies/µL for CT and NG respectively. Furthermore, it can successfully detect the DNA levels of these two bacteria without the need for DNA extraction and via a lateral flow-based platform. These assays thus hold the potential to be employed in clinics for rapid and efficient monitoring of sexually transmitted infections.
Keywords: Chlamydia trachomatis; Neisseria gonorrhoeae; gold nanoparticles; lateral flow assay; point-of-care; single-stranded oligonucleotides.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
Prof. Dipanjan Pan is the founder/cofounder of three University based start‐ups. None of these entities, however, supported this work.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
