Developing and Validating a Multivariable Prognostic-Predictive Classifier for Treatment Escalation of Oropharyngeal Squamous Cell Carcinoma: The PREDICTR-OPC Study
- PMID: 37870417
- PMCID: PMC10792360
- DOI: 10.1158/1078-0432.CCR-23-1013
Developing and Validating a Multivariable Prognostic-Predictive Classifier for Treatment Escalation of Oropharyngeal Squamous Cell Carcinoma: The PREDICTR-OPC Study
Abstract
Purpose: While there are several prognostic classifiers, to date, there are no validated predictive models that inform treatment selection for oropharyngeal squamous cell carcinoma (OPSCC).Our aim was to develop clinical and/or biomarker predictive models for patient outcome and treatment escalation for OPSCC.
Experimental design: We retrospectively collated clinical data and samples from a consecutive cohort of OPSCC cases treated with curative intent at ten secondary care centers in United Kingdom and Poland between 1999 and 2012. We constructed tissue microarrays, which were stained and scored for 10 biomarkers. We then undertook multivariable regression of eight clinical parameters and 10 biomarkers on a development cohort of 600 patients. Models were validated on an independent, retrospectively collected, 385-patient cohort.
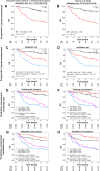
Results: A total of 985 subjects (median follow-up 5.03 years, range: 4.73-5.21 years) were included. The final biomarker classifier, comprising p16 and survivin immunohistochemistry, high-risk human papillomavirus (HPV) DNA in situ hybridization, and tumor-infiltrating lymphocytes, predicted benefit from combined surgery + adjuvant chemo/radiotherapy over primary chemoradiotherapy in the high-risk group [3-year overall survival (OS) 63.1% vs. 41.1%, respectively, HR = 0.32; 95% confidence interval (CI), 0.16-0.65; P = 0.002], but not in the low-risk group (HR = 0.4; 95% CI, 0.14-1.24; P = 0.114). On further adjustment by propensity scores, the adjusted HR in the high-risk group was 0.34, 95% CI = 0.17-0.67, P = 0.002, and in the low-risk group HR was 0.5, 95% CI = 0.1-2.38, P = 0.384. The concordance index was 0.73.
Conclusions: We have developed a prognostic classifier, which also appears to demonstrate moderate predictive ability. External validation in a prospective setting is now underway to confirm this and prepare for clinical adoption.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures



References
-
- Louie KS, Mehanna H, Sasieni P. Trends in head and neck cancers in England from 1995 to 2011 and projections up to 2025. Oral Oncol 2015;51:341–8. - PubMed
-
- Mehanna H, Beech T, Nicholson T, El-Hariry I, McConkey C, Paleri V, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta-analysis of trends by time and region. Head Neck 2013;35:747–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

