Testing and Management of Iron Overload After Genetic Screening-Identified Hemochromatosis
- PMID: 37870835
- PMCID: PMC10594145
- DOI: 10.1001/jamanetworkopen.2023.38995
Testing and Management of Iron Overload After Genetic Screening-Identified Hemochromatosis
Abstract
Importance: HFE gene-associated hereditary hemochromatosis type 1 (HH1) is underdiagnosed, resulting in missed opportunities for preventing morbidity and mortality.
Objective: To assess whether screening for p.Cys282Tyr homozygosity is associated with recognition and management of asymptomatic iron overload.
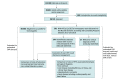
Design, setting, and participants: This cross-sectional study obtained data from the Geisinger MyCode Community Health Initiative, a biobank of biological samples and linked electronic health record data from a rural, integrated health care system. Participants included those who received a p.Cys282Tyr homozygous result via genomic screening (MyCode identified), had previously diagnosed HH1 (clinically identified), and those negative for p.Cys282Tyr homozygosity between 2017 and 2018. Data were analyzed from April 2020 to August 2023.
Exposure: Disclosure of a p.Cys282Tyr homozygous result.
Main outcomes and measures: Postdisclosure management and HFE-associated phenotypes in MyCode-identified participants were analyzed. Rates of HFE-associated phenotypes in MyCode-identified participants were compared with those of clinically identified participants. Relevant laboratory values and rates of laboratory iron overload among participants negative for p.Cys282Tyr homozygosity were compared with those of MyCode-identified participants.
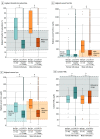
Results: A total of 86 601 participants had available exome sequences at the time of analysis, of whom 52 994 (61.4%) were assigned female at birth, and the median (IQR) age was 62.0 (47.0-73.0) years. HFE p.Cys282Tyr homozygosity was disclosed to 201 participants, of whom 57 (28.4%) had a prior clinical HH1 diagnosis, leaving 144 participants who learned of their status through screening. There were 86 300 individuals negative for p.Cys282Tyr homozygosity. After result disclosure, among MyCode-identified participants, 99 (68.8%) had a recommended laboratory test and 36 (69.2%) with laboratory or liver biopsy evidence of iron overload began phlebotomy or chelation. Fifty-three (36.8%) had iron overload; rates of laboratory iron overload were higher in MyCode-identified participants than participants negative for p.Cys282Tyr homozygosity (females: 34.1% vs 2.1%, P < .001; males: 39.0% vs 2.9%, P < .001). Iron overload (females: 34.1% vs 79.3%, P < .001; males: 40.7% vs 67.9%, P = .02) and some liver-associated phenotypes were observed at lower frequencies in MyCode-identified participants compared with clinically identified individuals.
Conclusions and relevance: Results of this cross-sectional study showed the ability of genomic screening to identify undiagnosed iron overload and encourage relevant management, suggesting the potential benefit of population screening for HFE p.Cys282Tyr homozygosity. Further studies are needed to examine the implications of genomic screening for health outcomes and cost-effectiveness.
Conflict of interest statement
Figures
References
-
- Online Mendelian Inheritance in Man (OMIM). Johns Hopkins University; 2016. September 8, 2016. Accessed June 7, 2021. https://www.omim.org/entry/235200
-
- GenBank . National Library of Medicine, National Center for Biotechnology Information. Accession No. NM_000410.4, Homo sapiens homeostatic iron regulator (HFE), transcript variant 1, mRNA. Accessed June 18, 2021. https://www.ncbi.nlm.nih.gov/nuccore/NM_000410
-
- Online Mendelian Inheritance in Man (OMIM). Johns Hopkins University; 2022. May 19, 2022. Accessed February 20, 2022. https://www.omim.org/entry/613609
-
- ClinVar . National Library of Medicine, National Center for Biotechnology Information. VCV000000009.24. Accessed June 7, 2021. https://www.ncbi.nlm.nih.gov/clinvar/variation/9/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

