Tissue Forge: Interactive biological and biophysics simulation environment
- PMID: 37871133
- PMCID: PMC10621971
- DOI: 10.1371/journal.pcbi.1010768
Tissue Forge: Interactive biological and biophysics simulation environment
Abstract
Tissue Forge is an open-source interactive environment for particle-based physics, chemistry and biology modeling and simulation. Tissue Forge allows users to create, simulate and explore models and virtual experiments based on soft condensed matter physics at multiple scales, from the molecular to the multicellular, using a simple, consistent interface. While Tissue Forge is designed to simplify solving problems in complex subcellular, cellular and tissue biophysics, it supports applications ranging from classic molecular dynamics to agent-based multicellular systems with dynamic populations. Tissue Forge users can build and interact with models and simulations in real-time and change simulation details during execution, or execute simulations off-screen and/or remotely in high-performance computing environments. Tissue Forge provides a growing library of built-in model components along with support for user-specified models during the development and application of custom, agent-based models. Tissue Forge includes an extensive Python API for model and simulation specification via Python scripts, an IPython console and a Jupyter Notebook, as well as C and C++ APIs for integrated applications with other software tools. Tissue Forge supports installations on 64-bit Windows, Linux and MacOS systems and is available for local installation via conda.
Copyright: © 2023 Sego et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
No competing interests.
Figures



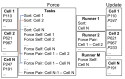

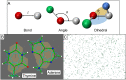



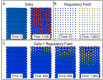
References
-
- Swat M., Thomas G., Belmonte J., Shirinifard A., Hmeljak D. & Glazier J. Multi-Scale Modeling of Tissues Using CompuCell3D. Methods In Cell Biology. 110 pp. 325–366 (2012), https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3612985/ doi: 10.1016/B978-0-12-388403-9.00013-8 - DOI - PMC - PubMed
-
- Graner F. & Glazier J. Simulation of biological cell sorting using a two-dimensional extended Potts model. Physical Review Letters. 69, 2013–2016 (1992,9), https://link.aps.org/doi/10.1103/PhysRevLett.69.2013, Publisher: American Physical Society; - DOI - PubMed
-
- Ghaffarizadeh A., Heiland R., Friedman S., Mumenthaler S. & Macklin P. PhysiCell: An open source physics-based cell simulator for 3-D multicellular systems. PLOS Computational Biology. 14, e1005991 (2018,2), https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1..., Publisher: Public Library of Science: - PMC - PubMed
-
- Mirams G., Arthurs C., Bernabeu M., Bordas R., Cooper J., Corrias A., Davit Y., Dunn S., Fletcher A., Harvey D., Marsh M., Osborne J., Pathmanathan P., Pitt-Francis J., Southern J., Zemzemi N. & Gavaghan D. Chaste: An Open Source C++ Library for Computational Physiology and Biology. PLOS Computational Biology. 9, e1002970 (2013,3), https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1..., Publisher: Public Library of Science; - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

