Neutralization escape by SARS-CoV-2 Omicron subvariant BA.2.86
- PMID: 37872011
- PMCID: PMC10842519
- DOI: 10.1016/j.vaccine.2023.10.051
Neutralization escape by SARS-CoV-2 Omicron subvariant BA.2.86
Abstract
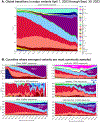
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) variant BA.2.86 has over 30 mutations in spike compared with BA.2 and XBB.1.5, which raised the possibility that BA.2.86 might evade neutralizing antibodies (NAbs) induced by vaccination or infection. In this study, we show that NAb titers are substantially lower to BA.2.86 compared with BA.2 but are similar or slightly higher than to other current circulating variants, including XBB.1.5, EG.5.1, and FL.1.5.1. Moreover, NAb titers against all these variants were higher in vaccinated individuals with a history of XBB.1.5 infection compared with vaccinated individuals with no history of XBB.1.5 infection, suggesting the potential utility of the monovalent XBB.1.5 mRNA boosters.
Keywords: BA.2.86; Neutralizing antibody; SARS-CoV-2; Vaccine; mRNA.
Copyright © 2023 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Dan Barouch reports financial support was provided by National Cancer Institute. Dan Barouch reports financial support was provided by Massachusetts Consortium on Pathogen Readiness. Dan Barouch reports financial support was provided by Ragon Institute.
Figures


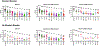

References
-
- Wang Q, Guo Y, Liu L, et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. bioRxiv 2023:2023.09.24.559214. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

