A genomic appraisal of invasive Salmonella Typhimurium and associated antibiotic resistance in sub-Saharan Africa
- PMID: 37872141
- PMCID: PMC10593746
- DOI: 10.1038/s41467-023-41152-6
A genomic appraisal of invasive Salmonella Typhimurium and associated antibiotic resistance in sub-Saharan Africa
Abstract
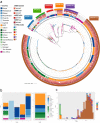
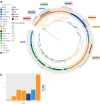
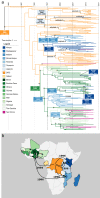
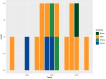
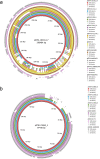
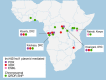
Invasive non-typhoidal Salmonella (iNTS) disease manifesting as bloodstream infection with high mortality is responsible for a huge public health burden in sub-Saharan Africa. Salmonella enterica serovar Typhimurium (S. Typhimurium) is the main cause of iNTS disease in Africa. By analysing whole genome sequence data from 1303 S. Typhimurium isolates originating from 19 African countries and isolated between 1979 and 2017, here we show a thorough scaled appraisal of the population structure of iNTS disease caused by S. Typhimurium across many of Africa's most impacted countries. At least six invasive S. Typhimurium clades have already emerged, with ST313 lineage 2 or ST313-L2 driving the current pandemic. ST313-L2 likely emerged in the Democratic Republic of Congo around 1980 and further spread in the mid 1990s. We observed plasmid-borne as well as chromosomally encoded fluoroquinolone resistance underlying emergences of extensive-drug and pan-drug resistance. Our work provides an overview of the evolution of invasive S. Typhimurium disease, and can be exploited to target control measures.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- (IHME) IfHMaE. in GBD 2019 Cause and Risk Summary: Invasive non-typhoidal Salmonella (iNTS). (eds) (Global Health Metrics, 2021). https://www.healthdata.org/results/gbd_summaries/2019/invasive-non-typho....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

