Phase separation of BuGZ regulates gut regeneration and aging through interaction with m6A regulators
- PMID: 37872148
- PMCID: PMC10593810
- DOI: 10.1038/s41467-023-42474-1
Phase separation of BuGZ regulates gut regeneration and aging through interaction with m6A regulators
Abstract
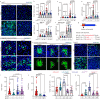
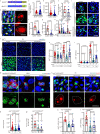
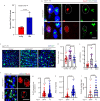
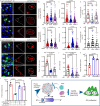
Exploring the role of phase separation in intracellular compartment formation is an active area of research. However, the associations of phase separation with intestinal stem cell (ISC)-dependent regeneration and aging remain unclear. Here, we demonstrate that BuGZ, a coacervating mitotic effector, shows age- and injury-associated condensation in Drosophila ISC nuclei during interphase. BuGZ condensation promotes ISC proliferation, affecting Drosophila gut repair and longevity. Moreover, m6A reader YT521-B acts as the transcriptional and functional downstream of BuGZ. The binding of YT521-B promotor or m6A writer Ime4/ Mettl14 to BuGZ controls its coacervation, indicating that the promotor may accelerate the phase transition of its binding transcription factor. Hence, we propose that phase separation and m6A regulators may be critical for ameliorating ISC-dependent gut regeneration and aging and requires further study.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

