HuR-mediated nucleocytoplasmic translocation of HOTAIR relieves its inhibition of osteogenic differentiation and promotes bone formation
- PMID: 37872163
- PMCID: PMC10593784
- DOI: 10.1038/s41413-023-00289-2
HuR-mediated nucleocytoplasmic translocation of HOTAIR relieves its inhibition of osteogenic differentiation and promotes bone formation
Abstract
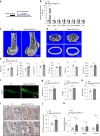
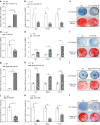
Bone marrow mesenchymal stem cell (BMSC) osteogenic differentiation and osteoblast function play critical roles in bone formation, which is a highly regulated process. Long noncoding RNAs (lncRNAs) perform diverse functions in a variety of biological processes, including BMSC osteogenic differentiation. Although several studies have reported that HOX transcript antisense RNA (HOTAIR) is involved in BMSC osteogenic differentiation, its effect on bone formation in vivo remains unclear. Here, by constructing transgenic mice with BMSC (Prx1-HOTAIR)- and osteoblast (Bglap-HOTAIR)-specific overexpression of HOTAIR, we found that Prx1-HOTAIR and Bglap-HOTAIR transgenic mice show different bone phenotypes in vivo. Specifically, Prx1-HOTAIR mice showed delayed bone formation, while Bglap-HOTAIR mice showed increased bone formation. HOTAIR inhibits BMSC osteogenic differentiation but promotes osteoblast function in vitro. Furthermore, we identified that HOTAIR is mainly located in the nucleus of BMSCs and in the cytoplasm of osteoblasts. HOTAIR displays a nucleocytoplasmic translocation pattern during BMSC osteogenic differentiation. We first identified that the RNA-binding protein human antigen R (HuR) is responsible for HOTAIR nucleocytoplasmic translocation. HOTAIR is essential for osteoblast function, and cytoplasmic HOTAIR binds to miR-214 and acts as a ceRNA to increase Atf4 protein levels and osteoblast function. Bglap-HOTAIR mice, but not Prx1-HOTAIR mice, showed alleviation of bone loss induced by unloading. This study reveals the importance of temporal and spatial regulation of HOTAIR in BMSC osteogenic differentiation and bone formation, which provides new insights into precise regulation as a target for bone loss.
© 2023. West China School of Stomatology Sichuan University.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 2011;13:27–38. - PubMed
-
- Franceschi RT, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Ann. N. Y. Acad. Sci. 2007;1116:196–207. - PubMed
-
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

