Beyond gene-disease validity: capturing structured data on inheritance, allelic requirement, disease-relevant variant classes, and disease mechanism for inherited cardiac conditions
- PMID: 37872640
- PMCID: PMC10594882
- DOI: 10.1186/s13073-023-01246-8
Beyond gene-disease validity: capturing structured data on inheritance, allelic requirement, disease-relevant variant classes, and disease mechanism for inherited cardiac conditions
Abstract
Background: As the availability of genomic testing grows, variant interpretation will increasingly be performed by genomic generalists, rather than domain-specific experts. Demand is rising for laboratories to accurately classify variants in inherited cardiac condition (ICC) genes, including secondary findings.
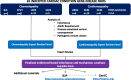
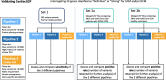
Methods: We analyse evidence for inheritance patterns, allelic requirement, disease mechanism and disease-relevant variant classes for 65 ClinGen-curated ICC gene-disease pairs. We present this information for the first time in a structured dataset, CardiacG2P, and assess application in genomic variant filtering.
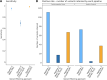
Results: For 36/65 gene-disease pairs, loss of function is not an established disease mechanism, and protein truncating variants are not known to be pathogenic. Using the CardiacG2P dataset as an initial variant filter allows for efficient variant prioritisation whilst maintaining a high sensitivity for retaining pathogenic variants compared with two other variant filtering approaches.
Conclusions: Access to evidence-based structured data representing disease mechanism and allelic requirement aids variant filtering and analysis and is a pre-requisite for scalable genomic testing.
Keywords: Allelic requirement; Disease mechanism; Gene curation; Genomic variant filtering; Inheritance; Inherited cardiac conditions; Variant classification; Variant interpretation.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
EMM is a Consultant for Amgen, AstraZeneca, Avidity Biosciences, Cytokinetics, PepGen, Pfizer, Stealth Biotherapeutics, and Tenaya Therapeutics and founder of Ikaika Therapeutics. CJ is a Consultant for Pfizer Inc (paid), StrideBio Inc (unpaid), and Tenaya Inc (unpaid). TL has research grant support from Pfizer. DPJ is a Consultant for Alexion, Alleviant, Cytokinetics, Novo Nordisk, Pfizer, and Tenaya Therapeutics. JI has research grant support from Bristol Myers Squibb. JSW has received research support or consultancy fees from Myokardia, Bristol-Myers Squibb, Pfizer, and Foresite Labs. The other authors declare that they have no competing interests.
Figures
Update of
-
Beyond gene-disease validity: capturing structured data on inheritance, allelic-requirement, disease-relevant variant classes, and disease mechanism for inherited cardiac conditions.medRxiv [Preprint]. 2023 Apr 3:2023.04.03.23287612. doi: 10.1101/2023.04.03.23287612. medRxiv. 2023. Update in: Genome Med. 2023 Oct 23;15(1):86. doi: 10.1186/s13073-023-01246-8. PMID: 37066275 Free PMC article. Updated. Preprint.
References
-
- Wilde AAM, Semsarian C, Márquez MF, Sepehri Shamloo A, Ackerman MJ, Ashley EA, et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the State of Genetic Testing for Cardiac Diseases. Heart Rhythm. 2022;19(7):e1–e60. doi: 10.1016/J.HRTHM.2022.03.1225. - DOI - PubMed
-
- Landstrom AP, Chahal AA, Ackerman MJ, Cresci S, Milewicz DM, Morris AA, et al. Interpreting incidentally identified variants in genes associated with heritable cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2023;16(2):E000092. doi: 10.1161/HCG.0000000000000092. - DOI - PubMed
-
- Miller DT, Lee K, Abul-Husn NS, Amendola LM, Brothers K, Chung WK, et al. ACMG SF v3.1 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2022;24(7):1407–1414. doi: 10.1016/j.gim.2022.04.006. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

