This is a preprint.
APOLLO: A genome-scale metabolic reconstruction resource of 247,092 diverse human microbes spanning multiple continents, age groups, and body sites
- PMID: 37873072
- PMCID: PMC10592896
- DOI: 10.1101/2023.10.02.560573
APOLLO: A genome-scale metabolic reconstruction resource of 247,092 diverse human microbes spanning multiple continents, age groups, and body sites
Update in
-
A genome-scale metabolic reconstruction resource of 247,092 diverse human microbes spanning multiple continents, age groups, and body sites.Cell Syst. 2025 Feb 19;16(2):101196. doi: 10.1016/j.cels.2025.101196. Epub 2025 Feb 12. Cell Syst. 2025. PMID: 39947184
Abstract
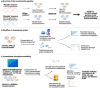
Computational modelling of microbiome metabolism has proved instrumental to catalyse our understanding of diet-host-microbiome-disease interactions through the interrogation of mechanistic, strain- and molecule-resolved metabolic models. We present APOLLO, a resource of 247,092 human microbial genome-scale metabolic reconstructions spanning 19 phyla and accounting for microbial genomes from 34 countries, all age groups, and five body sites. We explored the metabolic potential of the reconstructed strains and developed a machine learning classifier able to predict with high accuracy the taxonomic strain assignments. We also built 14,451 sample-specific microbial community models, which could be stratified by body site, age, and disease states. Finally, we predicted faecal metabolites enriched or depleted in gut microbiomes of people with Crohn's disease, Parkinson disease, and undernourished children. APOLLO is compatible with the human whole-body models, and thus, provide unprecedented opportunities for systems-level modelling of personalised host-microbiome co-metabolism. APOLLO will be freely available under https://www.vmh.life/.
Conflict of interest statement
Declaration of interests The authors declare no conflict of interest.
Figures




References
-
- Aden K., Rehman A., Waschina S., Pan W.H., Walker A., Lucio M., Nunez A.M., Bharti R., Zimmerman J., Bethge J., et al. (2019). Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology 157, 1279–1292 e1211. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
