This is a preprint.
The genetic basis of novel trait gain in walking fish
- PMID: 37873105
- PMCID: PMC10592820
- DOI: 10.1101/2023.10.14.562356
The genetic basis of novel trait gain in walking fish
Update in
-
Ancient developmental genes underlie evolutionary novelties in walking fish.Curr Biol. 2024 Oct 7;34(19):4339-4348.e6. doi: 10.1016/j.cub.2024.08.042. Epub 2024 Sep 26. Curr Biol. 2024. PMID: 39332403 Free PMC article.
Abstract
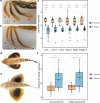
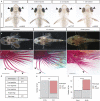
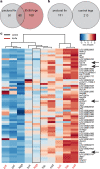
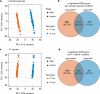
A major goal in biology is to understand how organisms evolve novel traits. Multiple studies have identified genes contributing to regressive evolution, the loss of structures that existed in a recent ancestor. However, fewer examples exist for genes underlying constructive evolution, the gain of novel structures and capabilities in lineages that previously lacked them. Sea robins are fish that have evolved enlarged pectoral fins, six mobile locomotory fin rays (legs) and six novel macroscopic lobes in the central nervous system (CNS) that innervate the corresponding legs. Here, we establish successful husbandry and use a combination of transcriptomics, CRISPR-Cas9 editing, and behavioral assays to identify key transcription factors that are required for leg formation and function in sea robins. We also generate hybrids between two sea robin species with distinct leg morphologies and use allele-specific expression analysis and gene editing to explore the genetic basis of species-specific trait diversity, including a novel sensory gain of function. Collectively, our study establishes sea robins as a new model for studying the genetic basis of novel organ formation, and demonstrates a crucial role for the conserved limb gene tbx3a in the evolution of chemosensory legs in walking fish.
Figures








Similar articles
-
Ancient developmental genes underlie evolutionary novelties in walking fish.Curr Biol. 2024 Oct 7;34(19):4339-4348.e6. doi: 10.1016/j.cub.2024.08.042. Epub 2024 Sep 26. Curr Biol. 2024. PMID: 39332403 Free PMC article.
-
Walking on chains: the morphology and mechanics behind the fin ray derived limbs of sea-robins.J Exp Biol. 2020 Sep 28;223(Pt 18):jeb227140. doi: 10.1242/jeb.227140. J Exp Biol. 2020. PMID: 32709626
-
Evolution of novel sensory organs in fish with legs.Curr Biol. 2024 Oct 7;34(19):4349-4356.e7. doi: 10.1016/j.cub.2024.08.014. Epub 2024 Sep 26. Curr Biol. 2024. PMID: 39332400 Free PMC article.
-
The Water to Land Transition Submerged: Multifunctional Design of Pectoral Fins for Use in Swimming and in Association with Underwater Substrate.Integr Comp Biol. 2022 Oct 29;62(4):908-921. doi: 10.1093/icb/icac061. Integr Comp Biol. 2022. PMID: 35652788 Free PMC article. Review.
-
Utilizing the blind cavefish Astyanax mexicanus to understand the genetic basis of behavioral evolution.J Exp Biol. 2020 Feb 7;223(Pt Suppl 1):jeb208835. doi: 10.1242/jeb.208835. J Exp Biol. 2020. PMID: 32034044 Review.
References
-
- Leal F. & Cohn M. J. Loss and re-emergence of legs in snakes by modular evolution of sonic hedgehog and HOXD enhancers. Curr. Biol. 26, 2966–2973 (2016). - PubMed
-
- Colosimo P. F., Hosemann K. E., Balabhadra S., Jr G. V., Dickson M., Grimwood J., Schmutz J., Myers R. M., Schluter D. & Kingsley D. M. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307, 1928–1933 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
