This is a preprint.
DengueSeq: A pan-serotype whole genome amplicon sequencing protocol for dengue virus
- PMID: 37873191
- PMCID: PMC10592998
- DOI: 10.1101/2023.10.13.23296997
DengueSeq: A pan-serotype whole genome amplicon sequencing protocol for dengue virus
Update in
-
DengueSeq: a pan-serotype whole genome amplicon sequencing protocol for dengue virus.BMC Genomics. 2024 May 1;25(1):433. doi: 10.1186/s12864-024-10350-x. BMC Genomics. 2024. PMID: 38693476 Free PMC article.
Abstract
Background: The increasing burden of dengue virus on public health due to more explosive and frequent outbreaks highlights the need for improved surveillance and control. Genomic surveillance of dengue virus not only provides important insights into the emergence and spread of genetically diverse serotypes and genotypes, but it is also critical to monitor the effectiveness of newly implemented control strategies. Here, we present DengueSeq, an amplicon sequencing protocol, which enables whole-genome sequencing of all four dengue virus serotypes.
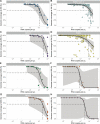
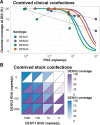
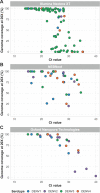
Results: We developed primer schemes for the four dengue virus serotypes, which can be combined into a pan-serotype approach. We validated both approaches using genetically diverse virus stocks and clinical specimens that contained a range of virus copies. High genome coverage (>95%) was achieved for all genotypes, except DENV2 (genotype VI) and DENV 4 (genotype IV) sylvatics, with similar performance of the serotype-specific and pan-serotype approaches. The limit of detection to reach 70% coverage was 101-102 RNA copies/μL for all four serotypes, which is similar to other commonly used primer schemes. DengueSeq facilitates the sequencing of samples without known serotypes, allows the detection of multiple serotypes in the same sample, and can be used with a variety of library prep kits and sequencing instruments.
Conclusions: DengueSeq was systematically evaluated with virus stocks and clinical specimens spanning the genetic diversity within each of the four dengue virus serotypes. The primer schemes can be plugged into existing amplicon sequencing workflows to facilitate the global need for expanded dengue virus genomic surveillance.
Keywords: Genomic surveillance; amplicon sequencing; dengue virus; next-generation sequencing; whole-genome sequencing.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures



Similar articles
-
DengueSeq: a pan-serotype whole genome amplicon sequencing protocol for dengue virus.BMC Genomics. 2024 May 1;25(1):433. doi: 10.1186/s12864-024-10350-x. BMC Genomics. 2024. PMID: 38693476 Free PMC article.
-
A Serotype-Specific and Multiplex PCR Method for Whole-Genome Sequencing of Dengue Virus Directly from Clinical Samples.Microbiol Spectr. 2022 Oct 26;10(5):e0121022. doi: 10.1128/spectrum.01210-22. Epub 2022 Sep 12. Microbiol Spectr. 2022. PMID: 36094197 Free PMC article.
-
DEN-IM: dengue virus genotyping from amplicon and shotgun metagenomic sequencing.Microb Genom. 2020 Mar;6(3):e000328. doi: 10.1099/mgen.0.000328. Microb Genom. 2020. PMID: 32134380 Free PMC article.
-
Detection of genotype-1 of dengue virus serotype 3 for the first time and complete genome analysis of dengue viruses during the 2018 epidemic in Mandalay, Upper Myanmar.PLoS One. 2021 Jun 4;16(6):e0251314. doi: 10.1371/journal.pone.0251314. eCollection 2021. PLoS One. 2021. PMID: 34086703 Free PMC article.
-
Emergence of dengue virus type 1 and type 3 as dominant serotypes during 2017 in Pune and Nashik regions of Maharashtra, Western India.Infect Genet Evol. 2018 Dec;66:272-283. doi: 10.1016/j.meegid.2018.10.016. Epub 2018 Oct 23. Infect Genet Evol. 2018. PMID: 30366083
References
-
- Dengue – the Region of the Americas [Internet]. [cited 2023 Jul 26]. Available from: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON475
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
