This is a preprint.
A Specialized Epithelial Cell Type Regulating Mucosal Immunity and Driving Human Crohn's Disease
- PMID: 37873404
- PMCID: PMC10592875
- DOI: 10.1101/2023.09.30.560293
A Specialized Epithelial Cell Type Regulating Mucosal Immunity and Driving Human Crohn's Disease
Update in
-
Identification and multimodal characterization of a specialized epithelial cell type associated with Crohn's disease.Nat Commun. 2024 Aug 22;15(1):7204. doi: 10.1038/s41467-024-51580-7. Nat Commun. 2024. PMID: 39169060 Free PMC article.
Abstract
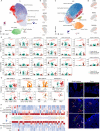
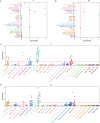
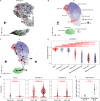
Crohn's disease (CD) is a complex chronic inflammatory disorder that may affect any part of gastrointestinal tract with extra-intestinal manifestations and associated immune dysregulation. To characterize heterogeneity in CD, we profiled single-cell transcriptomics of 170 samples from 65 CD patients and 18 non-inflammatory bowel disease (IBD) controls in both the terminal ileum (TI) and ascending colon (AC). Analysis of 202,359 cells identified a novel epithelial cell type in both TI and AC, featuring high expression of LCN2, NOS2, and DUOX2, and thus is named LND. LND cells, confirmed by high-resolution in-situ RNA imaging, were rarely found in non-IBD controls, but expanded significantly in active CD. Compared to other epithelial cells, genes defining LND cells were enriched in antimicrobial response and immunoregulation. Moreover, multiplexed protein imaging demonstrated that LND cell abundance was associated with immune infiltration. Cross-talk between LND and immune cells was explored by ligand-receptor interactions and further evidenced by their spatial colocalization. LND cells showed significant enrichment of expression specificity of IBD/CD susceptibility genes, revealing its role in immunopathogenesis of CD. Investigating lineage relationships of epithelial cells detected two LND cell subpopulations with different origins and developmental potential, early and late LND. The ratio of the late to early LND cells was related to anti-TNF response. These findings emphasize the pathogenic role of the specialized LND cell type in both Crohn's ileitis and Crohn's colitis and identify novel biomarkers associated with disease activity and treatment response.
Figures




References
-
- Arijs I., De Hertogh G., Lemaire K., Quintens R., Van Lommel L., Van Steen K., Leemans P., Cleynen I., Van Assche G., Vermeire S., et al. (2009). Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One 4, e7984. - PMC - PubMed
-
- Ayyaz A., Kumar S., Sangiorgi B., Ghoshal B., Gosio J., Ouladan S., Fink M., Barutcu S., Trcka D., Shen J., et al. (2019). Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125. - PubMed
-
- Badolato R., Wang J.M., Murphy W.J., Lloyd A.R., Michiel D.F., Bausserman L.L., Kelvin D.J., and Oppenheim J.J. (1994). Serum amyloid A is a chemoattractant: induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J Exp Med 180, 203–209. - PMC - PubMed
-
- Banerjee A., Herring C.A., Chen B., Kim H., Simmons A.J., Southard-Smith A.N., Allaman M.M., White J.R., Macedonia M.C., McKinley E.T., et al. (2020). Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 159, 2101–2115 e2105. - PMC - PubMed
Publication types
Grants and funding
- UL1 TR000445/TR/NCATS NIH HHS/United States
- I01 CX002473/CX/CSRD VA/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- UM1 CA183727/CA/NCI NIH HHS/United States
- P01 CA229123/CA/NCI NIH HHS/United States
- P01 AI139449/AI/NIAID NIH HHS/United States
- P50 CA236733/CA/NCI NIH HHS/United States
- I01 BX004366/BX/BLRD VA/United States
- R01 DK103831/DK/NIDDK NIH HHS/United States
- U54 CA274367/CA/NCI NIH HHS/United States
- R01 DK128200/DK/NIDDK NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- I01 CX002171/CX/CSRD VA/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
