Stroma AReactive Invasion Front Areas (SARIFA) proves prognostic relevance in gastric carcinoma and is based on a tumor-adipocyte interaction indicating an altered immune response
- PMID: 37874427
- PMCID: PMC10761465
- DOI: 10.1007/s10120-023-01436-8
Stroma AReactive Invasion Front Areas (SARIFA) proves prognostic relevance in gastric carcinoma and is based on a tumor-adipocyte interaction indicating an altered immune response
Abstract
Background: Recently, we presented Stroma AReactive Invasion Front Areas (SARIFA) as a new histomorphologic negative prognostic biomarker in gastric cancer. It is defined as direct contact between tumor cells and fat cells. The aim of this study was to further elucidate the underlying genomic, transcriptional, and immunological mechanisms of the SARIFA phenomenon.
Methods: To address these questions, SARIFA was classified on H&E-stained tissue sections of three cohorts: an external cohort (n = 489, prognostic validation), the TCGA-STAD cohort (n = 194, genomic and transcriptomic analysis), and a local cohort (n = 60, digital spatial profiling (whole transcriptome) and double RNA in situ hybridization/immunostaining of cytokines).
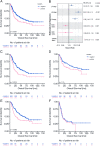
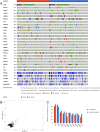
Results: SARIFA status proved to be an independent negative prognostic factor for overall survival in an external cohort of gastric carcinomas. In TCGA-STAD cohort, SARIFA is not driven by distinct genomic alterations, whereas the gene expression analyses showed an upregulation of FABP4 in SARIFA-positive tumors. In addition, the transcriptional regulations of white adipocyte differentiation, triglyceride metabolism, and catabolism were upregulated in pathway analyses. In the DSP analysis of SARIFA-positive tumors, FABP4 and the transcriptional regulation of white adipocyte differentiation were upregulated in macrophages. Additionally, a significantly lower expression of the cytokines IL6 and TNFα was observed at the invasion front.
Conclusions: SARIFA proves to be a strong negative prognostic biomarker in advanced gastric cancer, implicating an interaction of tumor cells with tumor-promoting adipocytes with crucial changes in tumor cell metabolism. SARIFA is not driven by tumor genetics but is very likely driven by an altered immune response as a causative mechanism.
Keywords: Biomarker; Gastric cancer; Histopathology; Molecular mechanisms; Stroma Areactive Invasion Front Areas (SARIFA).
© 2023. The Author(s).
Conflict of interest statement
BM has received compensations of travel expenses and fees for advisory board activities by Astra Zeneca, Boehringer Ingelheim, MERCK, MSD, BMS, Bayer, and Novartis, not related to this study. The other authors declare no potential conflicts of interest.
Figures




Comment on
-
Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer.Nat Med. 2023 Feb;29(2):430-439. doi: 10.1038/s41591-022-02134-1. Epub 2023 Jan 9. Nat Med. 2023. PMID: 36624314
References
-
- Martin B, Grosser B, Kempkens L, Miller S, Bauer S, Dhillon C, Banner BM, Brendel EM, Sipos E, Vlasenko D, et al. Stroma AReactive Invasion Front Areas (SARIFA)-a new easily to determine biomarker in colon cancer-results of a retrospective study. Cancers (Basel) 2021 doi: 10.3390/cancers13194880. - DOI - PMC - PubMed
-
- Grosser B, Gluckstein MI, Dhillon C, Schiele S, Dintner S, VanSchoiack A, Kroeppler D, Martin B, Probst A, Vlasenko D, et al. Stroma AReactive Invasion Front Areas (SARIFA) - a new prognostic biomarker in gastric cancer related to tumor-promoting adipocytes. J Pathol. 2022;256:71–82. doi: 10.1002/path.5810. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

