Targeting eIF4A triggers an interferon response to synergize with chemotherapy and suppress triple-negative breast cancer
- PMID: 37874652
- PMCID: PMC10721161
- DOI: 10.1172/JCI172503
Targeting eIF4A triggers an interferon response to synergize with chemotherapy and suppress triple-negative breast cancer
Abstract
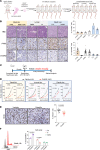
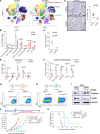
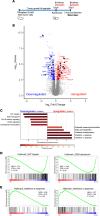
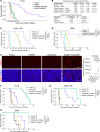
Protein synthesis is frequently dysregulated in cancer and selective inhibition of mRNA translation represents an attractive cancer therapy. Here, we show that therapeutically targeting the RNA helicase eIF4A with zotatifin, the first-in-class eIF4A inhibitor, exerts pleiotropic effects on both tumor cells and the tumor immune microenvironment in a diverse cohort of syngeneic triple-negative breast cancer (TNBC) mouse models. Zotatifin not only suppresses tumor cell proliferation but also directly repolarizes macrophages toward an M1-like phenotype and inhibits neutrophil infiltration, which sensitizes tumors to immune checkpoint blockade. Mechanistic studies revealed that zotatifin reprograms the tumor translational landscape, inhibits the translation of Sox4 and Fgfr1, and induces an interferon (IFN) response uniformly across models. The induction of an IFN response is partially due to the inhibition of Sox4 translation by zotatifin. A similar induction of IFN-stimulated genes was observed in breast cancer patient biopsies following zotatifin treatment. Surprisingly, zotatifin significantly synergizes with carboplatin to trigger DNA damage and an even heightened IFN response, resulting in T cell-dependent tumor suppression. These studies identified a vulnerability of eIF4A in TNBC, potential pharmacodynamic biomarkers for zotatifin, and provide a rationale for new combination regimens consisting of zotatifin and chemotherapy or immunotherapy as treatments for TNBC.
Keywords: Breast cancer; Immunotherapy; Oncology; Translation.
Conflict of interest statement
Figures



Update of
-
Targeting EIF4A triggers an interferon response to synergize with chemotherapy and suppress triple-negative breast cancer.bioRxiv [Preprint]. 2023 Sep 28:2023.09.28.559973. doi: 10.1101/2023.09.28.559973. bioRxiv. 2023. Update in: J Clin Invest. 2023 Dec 15;133(24):e172503. doi: 10.1172/JCI172503. PMID: 37808840 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

