The groundnut improvement network for Africa (GINA) germplasm collection: a unique genetic resource for breeding and gene discovery
- PMID: 37875136
- PMCID: PMC10755195
- DOI: 10.1093/g3journal/jkad244
The groundnut improvement network for Africa (GINA) germplasm collection: a unique genetic resource for breeding and gene discovery
Abstract
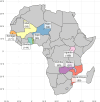
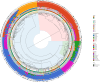
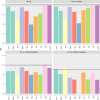
Cultivated peanut or groundnut (Arachis hypogaea L.) is a grain legume grown in many developing countries by smallholder farmers for food, feed, and/or income. The speciation of the cultivated species, that involved polyploidization followed by domestication, greatly reduced its variability at the DNA level. Mobilizing peanut diversity is a prerequisite for any breeding program for overcoming the main constraints that plague production and for increasing yield in farmer fields. In this study, the Groundnut Improvement Network for Africa assembled a collection of 1,049 peanut breeding lines, varieties, and landraces from 9 countries in Africa. The collection was genotyped with the Axiom_Arachis2 48K SNP array and 8,229 polymorphic single nucleotide polymorphism (SNP) markers were used to analyze the genetic structure of this collection and quantify the level of genetic diversity in each breeding program. A supervised model was developed using dapc to unambiguously assign 542, 35, and 172 genotypes to the Spanish, Valencia, and Virginia market types, respectively. Distance-based clustering of the collection showed a clear grouping structure according to subspecies and market types, with 73% of the genotypes classified as fastigiata and 27% as hypogaea subspecies. Using STRUCTURE, the global structuration was confirmed and showed that, at a minimum membership of 0.8, 76% of the varieties that were not assigned by dapc were actually admixed. This was particularly the case of most of the genotype of the Valencia subgroup that exhibited admixed genetic heritage. The results also showed that the geographic origin (i.e. East, Southern, and West Africa) did not strongly explain the genetic structure. The gene diversity managed by each breeding program, measured by the expected heterozygosity, ranged from 0.25 to 0.39, with the Niger breeding program having the lowest diversity mainly because only lines that belong to the fastigiata subspecies are used in this program. Finally, we developed a core collection composed of 300 accessions based on breeding traits and genetic diversity. This collection, which is composed of 205 genotypes of fastigiata subspecies (158 Spanish and 47 Valencia) and 95 genotypes of hypogaea subspecies (all Virginia), improves the genetic diversity of each individual breeding program and is, therefore, a unique resource for allele mining and breeding.
Keywords: breeding; core collection; genotyping; germplasm diversity; groundnut improvement; network.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures
References
-
- Achola E, Wasswa P, Fonceka D, Clevenger JP, Bajaj P, Ozias-Akins P, Rami J-F, Deom CM, Hoisington DA, Edema R, et al. 2023. Genome-wide association studies reveal novel loci for resistance to groundnut rosette disease in the African core groundnut collection. Theor Appl Genet. 136(3):35. doi:10.1007/s00122-023-04259-4. - DOI - PMC - PubMed
-
- Barrandeguy ME, García MV. 2021. The sensitiveness of expected heterozygosity and allelic richness estimates for analyzing population genetic diversity. In: Trindade Maia R, de Araújo Campos M, editors. Genetic Variation. IntechOpen.
-
- Bertioli DJ, Clevenger J, Godoy IJ, Stalker HT, Wood S, Santos JF, Ballén-Taborda C, Abernathy B, Azevedo V, Campbell J, et al. 2021. Legacy genetics of Arachis cardenasii in the peanut crop shows the profound benefits of international seed exchange. Proc Natal Acad Sci U S A. 118(38):e2104899118. doi:10.1073/pnas.2104899118. - DOI - PMC - PubMed
-
- Bertioli DJ, Seijo G, Freitas FO, Valls JFM, Leal-Bertioli SCM, Moretzsohn MC. 2011. An overview of peanut and its wild relatives. Plant Genet Resour. 9(01):134–149. doi:10.1017/S1479262110000444. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
