Integrated radiogenomics models predict response to neoadjuvant chemotherapy in high grade serous ovarian cancer
- PMID: 37875466
- PMCID: PMC10598212
- DOI: 10.1038/s41467-023-41820-7
Integrated radiogenomics models predict response to neoadjuvant chemotherapy in high grade serous ovarian cancer
Abstract
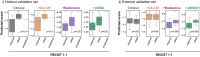
High grade serous ovarian carcinoma (HGSOC) is a highly heterogeneous disease that typically presents at an advanced, metastatic state. The multi-scale complexity of HGSOC is a major obstacle to predicting response to neoadjuvant chemotherapy (NACT) and understanding critical determinants of response. Here we present a framework to predict the response of HGSOC patients to NACT integrating baseline clinical, blood-based, and radiomic biomarkers extracted from all primary and metastatic lesions. We use an ensemble machine learning model trained to predict the change in total disease volume using data obtained at diagnosis (n = 72). The model is validated in an internal hold-out cohort (n = 20) and an independent external patient cohort (n = 42). In the external cohort the integrated radiomics model reduces the prediction error by 8% with respect to the clinical model, achieving an AUC of 0.78 for RECIST 1.1 classification compared to 0.47 for the clinical model. Our results emphasize the value of including radiomics data in integrative models of treatment response and provide methods for developing new biomarker-based clinical trials of NACT in HGSOC.
© 2023. Springer Nature Limited.
Conflict of interest statement
F.Mo., D.C., A.P., J.D.B. and N.R. are inventors of the patent “Enhanced detection of target DNA by fragment size analysis” (WO/2020/094775). N.R. and J.D.B. are inventors of the patent “TAm-Seq v2 method for ctDNA estimation” (WO 2016/009224A1). T.G. is an employee of and owns shares from AstraZeneca. F.Ma. and J.D.B. are founders and directors of Tailor Bio Ltd and own shares in Tailor Bio Ltd. J.D.B. has received honoraria and personal payments from AstraZeneca, GSK and Clovis Oncology. M.C.O. has received honoraria from GSK. E.S. is co-founder and shareholder of Lucida Medical Ltd. L.R. has received consulting fees from Lucida Medical Ltd. The remaining authors declare no competing interests.
Figures




References
-
- Vergote I, et al. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N. Engl. J. Med. 2010;363:943–953. - PubMed
-
- Kehoe S, et al. Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (CHORUS): An open-label, randomised, controlled, non-inferiority trial. Lancet. 2015;386:249–257. - PubMed
-
- Van Meurs HS, et al. Which patients benefit most from primary surgery or neoadjuvant chemotherapy in stage IIIC or IV ovarian cancer? An exploratory analysis of the European Organisation for Research and Treatment of Cancer 55971 randomised trial. Eur. J. Cancer. 2013;49:3191–3201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RG98755/WT_/Wellcome Trust/United Kingdom
- BRC-1215-20014/DH_/Department of Health/United Kingdom
- A19274/CRUK_/Cancer Research UK/United Kingdom
- A25117/CRUK_/Cancer Research UK/United Kingdom
- MRC_/Medical Research Council/United Kingdom
- 28292/CRUK_/Cancer Research UK/United Kingdom
- A17197/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- C9545/A29580/CRUK_/Cancer Research UK/United Kingdom
- A22905/CRUK_/Cancer Research UK/United Kingdom
- A20240/CRUK_/Cancer Research UK/United Kingdom
- A27066/CRUK_/Cancer Research UK/United Kingdom
- RC-1215-20014/DH_/Department of Health/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

