A window into the human immune system: comprehensive characterization of the complexity of antibody complementary-determining regions in functional antibodies
- PMID: 37876265
- PMCID: PMC10601506
- DOI: 10.1080/19420862.2023.2268255
A window into the human immune system: comprehensive characterization of the complexity of antibody complementary-determining regions in functional antibodies
Abstract
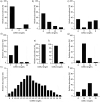
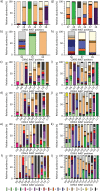
The human immune system uses antibodies to neutralize foreign antigens. They are composed of heavy and light chains, both with constant and variable regions. The variable region has six hypervariable loops, also known as complementary-determining regions (CDRs) that determine antibody diversity and antigen specificity. Knowledge of their significance, and certain residues present in these areas, is vital for antibody therapeutics development. This study includes an analysis of more than 11,000 human antibody sequences from the International Immunogenetics information system (IMGT). The analysis included parameters such as length distribution, overall amino acid diversity, amino acid frequency per CDR and residue position within antibody chains. Overall, our findings confirm existing knowledge, such as CDRH3's high length diversity and amino acid variability, increased aromatic residue usage, particularly tyrosine, charged and polar residues like aspartic acid, serine, and the flexible residue glycine. Specific residue positions within each CDR influence these occurrences, implying a unique amino acid type distribution pattern. We compared amino acid type usage in CDRs and non-CDR regions, both in globular and transmembrane proteins, which revealed distinguishing features, such as increased frequency of tyrosine, serine, aspartic acid, and arginine. These findings should prove useful for future optimization, improvement of affinity, synthetic antibody library design, or the creation of antibodies de-novo in silico.
Keywords: Amino acid diversity; Complementary-Determining Regions (CDRs); IMGT (International Immunogenetics Information System); antibodies/immunoglobulins; antibody sequencing; antibody therapeutics development; antibody-antigen complexes; de-novo in silico antibody design; immunology; synthetic antibody libraries.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
Amino acids characterization based on frequency and interaction analysis in human antigen-antibody complexes from Thera-SAbDab.Hum Antibodies. 2025 May;33(1-2):25-39. doi: 10.1177/10932607241303614. Epub 2025 Jan 23. Hum Antibodies. 2025. PMID: 39973811
-
High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries.J Mol Biol. 2007 Nov 2;373(4):924-40. doi: 10.1016/j.jmb.2007.08.005. Epub 2007 Aug 19. J Mol Biol. 2007. PMID: 17825836
-
Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions.J Mol Biol. 2004 Apr 23;338(2):299-310. doi: 10.1016/j.jmb.2004.02.050. J Mol Biol. 2004. PMID: 15066433
-
A potential antibody repertoire diversification mechanism through tyrosine sulfation for biotherapeutics engineering and production.Front Immunol. 2022 Dec 8;13:1072702. doi: 10.3389/fimmu.2022.1072702. eCollection 2022. Front Immunol. 2022. PMID: 36569848 Free PMC article.
-
Significance of antibody numbering systems in the development of antibody engineering.Hum Antibodies. 2023;31(4):71-80. doi: 10.3233/HAB-230014. Hum Antibodies. 2023. PMID: 38217590 Review.
Cited by
-
Amino Acids Frequency and Interaction Trends: Comprehensive Analysis of Experimentally Validated Viral Antigen-Antibody Complexes.Mol Biotechnol. 2025 Jan 8. doi: 10.1007/s12033-024-01361-w. Online ahead of print. Mol Biotechnol. 2025. PMID: 39775710
-
Benchmarking inverse folding models for antibody CDR sequence design.PLoS One. 2025 Jun 4;20(6):e0324566. doi: 10.1371/journal.pone.0324566. eCollection 2025. PLoS One. 2025. PMID: 40465588 Free PMC article.
-
Exploring Experimental and In Silico Approaches for Antibody-Drug Conjugates in Oncology Therapies.Pharmaceuticals (Basel). 2025 Aug 14;18(8):1198. doi: 10.3390/ph18081198. Pharmaceuticals (Basel). 2025. PMID: 40872592 Free PMC article. Review.
-
Identification of B-cell epitopes of Indian Zika virus strains using immunoinformatics.Front Immunol. 2025 Feb 27;16:1534737. doi: 10.3389/fimmu.2025.1534737. eCollection 2025. Front Immunol. 2025. PMID: 40083545 Free PMC article.
-
A synthetic heavy chain variable domain antibody library (VHL) provides highly functional antibodies with favorable developability.Protein Sci. 2025 Apr;34(4):e70090. doi: 10.1002/pro.70090. Protein Sci. 2025. PMID: 40100169
References
-
- Kienzler A-K, Eibel H.. Human B cell development and tolerance. In: Ratcliffe MJH, editor. Encyclopedia of immunobiology. United Kingdom: Elsevier; 2016. p. 105–12.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
