Ataluren improves myelopoiesis and neutrophil chemotaxis by restoring ribosome biogenesis and reducing p53 levels in Shwachman-Diamond syndrome cells
- PMID: 37876306
- PMCID: PMC10843527
- DOI: 10.1111/bjh.19134
Ataluren improves myelopoiesis and neutrophil chemotaxis by restoring ribosome biogenesis and reducing p53 levels in Shwachman-Diamond syndrome cells
Abstract
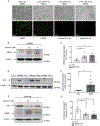
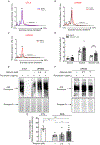
Shwachman-Diamond syndrome (SDS) is characterized by neutropenia, exocrine pancreatic insufficiency and skeletal abnormalities. SDS bone marrow haematopoietic progenitors show increased apoptosis and impairment in granulocytic differentiation. Loss of Shwachman-Bodian-Diamond syndrome (SBDS) expression results in reduced eukaryotic 80S ribosome maturation. Biallelic mutations in the SBDS gene are found in ~90% of SDS patients, ~55% of whom carry the c.183-184TA>CT nonsense mutation. Several translational readthrough-inducing drugs aimed at suppressing nonsense mutations have been developed. One of these, ataluren, has received approval in Europe for the treatment of Duchenne muscular dystrophy. We previously showed that ataluren can restore full-length SBDS protein synthesis in SDS-derived bone marrow cells. Here, we extend our preclinical study to assess the functional restoration of SBDS capabilities in vitro and ex vivo. Ataluren improved 80S ribosome assembly and total protein synthesis in SDS-derived cells, restored myelopoiesis in myeloid progenitors, improved neutrophil chemotaxis in vitro and reduced neutrophil dysplastic markers ex vivo. Ataluren also restored full-length SBDS synthesis in primary osteoblasts, suggesting that its beneficial role may go beyond the myeloid compartment. Altogether, our results strengthened the rationale for a Phase I/II clinical trial of ataluren in SDS patients who harbour the nonsense mutation.
Keywords: Shwachman-Diamond syndrome; ataluren; inherited bone marrow failure syndromes; myelodysplastic syndromes; translational readthrough-inducing drugs.
© 2023 The Authors. British Journal of Haematology published by British Society for Haematology and John Wiley & Sons Ltd.
Conflict of interest statement
Conflict of Interest Disclosures
V.B. and M.C. are co-inventors of the patent WO2018/050706 A1 Method of treatment of Shwachman-Diamond syndrome. The remaining authors declare no competing financial interests.
Figures



References
-
- Myers KC, Furutani E, Weller E, Siegele B, Galvin A, Arsenault V, et al. Clinical features and outcomes of patients with Shwachman-Diamond syndrome and myelodysplastic syndrome or acute myeloid leukaemia: a multicentre, retrospective, cohort study. Lancet Haematol. 2020;7(3):e238–e46. - PMC - PubMed
-
- Dror Y, Freedman MH. Shwachman-Diamond syndrome: An inherited preleukemic bone marrow failure disorder with aberrant hematopoietic progenitors and faulty marrow microenvironment. Blood. 1999;94(9):3048–54. - PubMed
-
- Donadieu J, Leblanc T, Bader Meunier B, Barkaoui M, Fenneteau O, Bertrand Y, et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90(1):45–53. - PubMed
-
- Elghetany MT, Alter BP. p53 protein overexpression in bone marrow biopsies of patients with Shwachman-Diamond syndrome has a prevalence similar to that of patients with refractory anemia. Arch Pathol Lab Med. 2002;126(4):452–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

